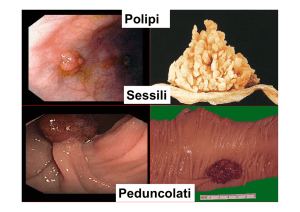
#
$ %
$&
' ( )&
*
!!"
Il cancro e’ una malattia genica, ovvero
!
insorge in conseguenza dell’accumulo
nella stessa cellula di alterazioni di
%
%
"
#
$ $
%
&
'
geni di diverse categorie
(oncogeni ed oncosoppressori)
!
#
$ $
%
&
*
%
$ (
))
Modello di Progressione del Tumore Colorettale
APC
APC
Met (p16)
APC
Met (p16)
K-RAS
APC
Met (p16)
K-RAS
DCC
APC
Met (p16)
K-RAS
DCC
p53
+ %
$%
,
'
(
%
+
-
Iper
proliferazione
Adenoma
tipo I
Adenoma
tipo II
Carcinoma
in situ
+
-
$
,
+
-$
(
&
,
-
,
Metastasi
1
'
'
(
(
'
%
)
)
%
)
2
)
' )
.
%
$-
+ 2 ,
-
4
-
3
/
/
%
/
.
.
-
Nel 5-8% dei casi il cancro e’ anche una
-
2
malattia genetica (una prima alterazione
'
(
%
)
)
(
(
)
'
genica e’ presente nella linea
1
2
$
/
-
germinale dell’individuo)
'
-
-
&
0
+0 #
-
(
-
.
$
,+
)
)
-
)
- $$
-
,
.
-
/
$
-
Familial Adenomatous Polyposis (FAP)
5
2
5
6
78
2
5 ;
(
97: 7 7
-<
(
" 7=
5
8
(
Linked families
!=
Cellular Localization
6
9
Function
1
The APC-β
β-catenin pathway
normal
cell
5q21
> 90%
Gene Product
2
<
77*
APC
(clonato nel 1991)
Chromosome
;9! =
5
5
Gene
E-cadherin
E-cadherin
tumoral
cell
cytoplasm
cell adhesion
Modello di Progressione del Tumore Colorettale
APC
P
GSK-3β
P
APC
P
proteasome
axin
Mutant
APC,
β -catenin,
axin
GSK-3β
APC
axin
P
P
degradation
CBP
groucho
CBP
groucho
Tcf
myc repressed
Tcf
myc activated
Iper
proliferazione
Adenoma
tipo I
Adenoma
tipo II
Carcinoma
in situ
Metastasi
2
Modello di Progressione del Tumore Colorettale
%
)
))
(
)
1
'
'
)
.
$ (
'
))
' .
1
.
'
)
%
'
(
(
'
' ) )
(
.
4 (
.
)
MYH
APC
(
0%
(
(
%7 )
(
'
2 / 0 /"
'
"
3
3
'
)
%
( .
.
%
8
(
)
#5
.
.
#5
'
( .
.
% ()
.
6&
( ) % 6&
9 /4
% ()
.
() %
Iper
proliferazione
Adenoma
tipo I
Adenoma
tipo II
Carcinoma
in situ
Metastasi
AMSTERDAM CRITERIA
(AC I)
1) At least three relatives with CRC, one of
whom must be a firstfirst-degree relative of
the other two. FAP must be excluded.
2) At least two generations must be
affected.
3) At least one individual must be less
than 50 years old at diagnosis.
!
)-
(
>
%
)
'%
Vasen et al.,
al., Dis Colon Rectum 1991
!!7
MECHANISM OF DNADNA-MISMATCH REPAIR IN E. COLI
Hereditary Non Polyposis Colorectal Cancer (HNPCC)
Gene
Chromosome
Linked families
MSH2
MSH1
2p21-22
3p21-23
60%
30%
Gene Product
Cellular Localization
Function
nucleus
repair of DNA
replication errors
3
ICG Definition of HNPCC
Mismatch Repair Genes
(MMR)
~
hMSH2 on chromosome 2p
~
hMLH1 on chromosome 3p
~
hPMS1 on chromosome 2q
~
hPMS2 on chromosome 7q
~
hMSH6 on chromosome 2p
• Familial clustering of CRC and/or endometrial ca.
• Associated cancer: ca. of the stomach, ovary, ureter/renal
pelvis, brain, small bowel, hepatobiliary tract and skin
• Development of cancer at an early stage
• Development of multiple cancers
• High history of MSI (MSI-H)
• Immunohistochemistry: loss of MLH1, MSH2 or
MSH6 protein expression
• Germline mutation in MMR genes
(MSH2, MLH1, MSH6, PMS1, PMS2)
Modello del Tumore Colorettale Sporadico
APC
APC
Met (p16)
Iper
proliferazione
Adenoma
tipo I
APC
Met (p16)
K-RAS
APC
Met (p16)
K-RAS
DCC
APC
Met (p16)
K-RAS
DCC
p53
Adenoma
tipo II
Carcinoma
in situ
Metastasi
Modello di Tumore Colorettale Ereditario
Mutazione germinale in APC
Met (p16)
Met (p16)
K-RAS
Adenoma
tipo I
Adenoma
tipo II
Growth
factor
APC
Met (p16)
Met (p16)
K-RAS
DCC
p53
Carcinoma
in situ
Metastasi
The RAS-RAF-MAPK signalling pathway
Modello di Progressione del Tumore Colorettale
APC
Met (p16)
K-RAS
DCC
OUT
APC
Met (p16)
K-RAS
P
P
B
GR
Tyrosine
Kinase
receptor
Cyclic
AMP
RAS
SOS
Growth
factor
Adenylate
cyclase
P
BRAF
P
G-protein
P
G-proteincoupled
receptor
P
MEK
ERK1 ERK2
P
Iper
proliferazione
Adenoma
tipo I
Adenoma
tipo II
Carcinoma
in situ
Metastasi
P
P
P
Cytoplasm
Changes in
gene expression
Nucleus
4
Modello di Progressione del Tumore Colorettale
ANALISI DI K-RAS PER PCR-RFLP
sequenza wt
ttggagctggtggcgtaggca
K-ras esone 1
5'
APC
Met (p16)
APC
Met (p16)
K-RAS
Iper
proliferazione
Adenoma
tipo I
Adenoma
tipo II
sequenza mutata
ttggagctgatggcgtaggca
APC
Met (p16)
K-RAS
DCC
Un primer modificato introduce un sito
di restrizione BstN I (in rosso)
ttggacctggtggcgtaggca
PCR
APC
3'
5'
3'
Il sito di restrizione BstN I non si crea
a causa della mutazione preesistente
ttggacctgatggcgtaggca
Digestione c on l’enzima di restrizione BstN I e separazione su gel di poliacrilammide al 7%.
wt
mut
140bp
111bp
Carcinoma
in situ
Metastasi
Modello di Progressione del Tumore Colorettale
APC
APC
Met (p16)
APC
Met (p16)
K-RAS
Iper
proliferazione
Adenoma
tipo I
Adenoma
tipo II
APC
Met (p16)
K-RAS
DCC
Carcinoma
in situ
APC
Met (p16)
K-RAS
DCC
p53
Metastasi
5
4
7
%
//
~ 50%
D 1
%
:
%
)
'
7+ ; $
)(
<
('
C
0
%
A
H
0
B=
=
"
2
G . 7:
2
G D 12
;
$
# .+
,
+ ,
"=
9=
( =
9=
/
$
9
=
G . 7:
@
2
-7:
7
?" ! =
--
9
~50%
--
"
~50%
--
=
-
--
-
:=
& $
- + ,
+ ,
/β
β/%
)
7+ ;
4
(
#2
! ! E * *F * B :B 9
1
! ! B ! FB ! 7
/β
β/%
'
)
COLON
*
T P 53 mut ,
C h 18 lo ss
19 %
K- R A S mut ,
T P 53 mut
9%
no mut
10 %
K- R A S mut
14 %
T P53 mut
9%
C h 18 lo ss
12 %
1
!!B
K- R A S mut ,
T P53 mut ,
C h18 lo ss
18 %
!FB! 7
7+ ;
C h 18 lo ss,
T P 53 mut no mut
16 %
7% K- R A S mut
K- R A S mut ,
16 %
T P53 mut
K- R A S mut ,
7%
C h 18 loss
11%
K- R A S mut ,
T P53 mut
C
h
18
lo
ss,
11%
TP53 mut
C h 18 lo ss
20%
12 %
K- R A S mut ,
C h 18 lo ss
9%
/
C h 18 l o ss,
T P 53 mut
28%
no mut
17%
*
T P 53 mut
7%
!!B
K- R A S mut
14 %
K- R A S mut ,
C h 18 l o s s
0%
K- R A S mut ,
T P5 3 mut
10 %
(
1
Frattini et al, Clin Cancer Res, 2004
RECTUM
% /
$
&
//
'
7"
C h 18 l o s s
14 %
K- R A S mut ,
C h 18 lo ss,
T P 53 mut
10 %
!FB! 7
6
!
5)
-
:
5 >12
1 /F 1 /
2 12
(
!
>12
/
5
6
(
5)
!
/
-
>12
-
>12
5 ;
7*9
>12
-
6
:
-
@
6
.
>12
0#
-
6
C 7**) +?* ! =
>12 6
+"7= , . +B7= ,
7
4
/
+ B= ,
/
!/
&
+
*
)
4
/
>
41
$
2
12
3
2
$
$
-
/
.$
D
G
0
>12
-
)D
)1D
>12
%
&
!= ,
-
$
7+ ;
3H.
/
$
=,
+: ! B ! = ,
+ -
>12
,
7(
(
!
$
/
! (
/
5
G
/
(
(
F 1
E 1:
E 1
>12
1
2
5
5
6
-
4G-proteincoupled
receptor
2
I
)1D
G
#
$
%-
"!
)%
'
.D
$
-
.D
D 1
1 J
-
#
>12
$ %
!! EB 9F* : B (
7
"!
"!
&
4 /
7
I )4 1
)4 1
%-
)4 1
)4 1
%-
D $ '2
%
&
&
9A* -
#
)
'
! ! : 9F
(
1
&
$-
)4 1
2.
$
/
A
7A79
-
)4 1
&
/
7=
I
/
#
-
&
H 2
' -
/
-
$-
/-
/
# )'
+
! ! B E: 7!F * : *
D 1
! ! B E: ! B F B* " 7! !
+
F +2
/
$
!!BE
I
!!B,
/
/
!!BE
! ! B ED
!! ,
$
$
$
! ! 7E 6
!!B,
! ! 7,
-
/
:!
)4 1
$
-
+
//%
!!BE
/ )4 1 %$ 0 ,-
$ 0
$-
&
&
+
)4 1
$
+
%
/ )4 1
%
#
9 (
%-
$ 0
)4 1
-
)4 1
.
/
B
"9
//
$
&
/
79
!!: *F
%
%
-
-
%
-
/
1# 2
4
"!
7!=
0
$
A
-
%
-
!! ,
/
%
+ 6
!! ,
=*
D: 2 ( 2
%
/
/
&
-
&
&
/
/
-
(.
/
&
(
! ! B E:! B F77B
8