RNA regolatori e controllo posttrascrizionale dell’espressione
genica
21 Aprile 2017
Corso di Genetica Molecolare
CeciliaMannironi
Is.tutodiBiologiaePatologiaMolecolaridelCNR
[email protected]
Human Genome Project (HGP)
1990-2003 Progetto di sequenziamento delle regioni eucromatiche del genoma. Non sono
state sequenziate le regioni eterocromatiche, dei centromeri e dei telomeri. In
parallelo sono stati sequenziati i genomi di vari organismi modello come
Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis ecc
Febbraio 2011
HumanGeneCount:MoreThanaChicken,LessThanaGrape
NB!inumerisonorela.viaigenicodifican.perproteine
PerteaM,SalzbergSL.2010.Betweenachickenandagrape:es.ma.ngthenumberofhumangenes.
GenomeBiol11:206.
La stima del numero dei geni umani si è ridotta progressivamente con lo
sviluppo di nuove tecnologie per il sequenziamento del DNA
MOLECULAR BIOLOGY AND EVOLUTION: Can Genes Explain Biological
Complexity?
Szathmary et al. Science 18 May 2001: Vol. 292. no. 5520, pp. 1315 – 1316
Il n° dei geni codificanti per proteine presenti nel genoma di un
organismo non è una misura della sua complessita’ biologica.
Le regioni codificanti del genoma sono identiche al 99.7% nell’uomo e
nello scimpanzè
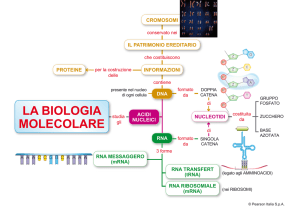
I COMPONENTI DEL GENOMA UMANO
retrotransposons
TRASCRIZIONE PERVASIVA
Il 70% del genoma umano è trascritto ma solo l’1.5-2% dei trascritti
codifica per proteine
COMPOSIZIONE DEL TRASCRITTOMA NEI MAMMIFERI
Gli RNA non codificanti (non coding RNAs, ncRNAs) sono trascritti che non
contengono open reading frames (ORF) o che hanno accumulato mutazione
che li rendono inattivi (pseudogeni). Caratteristiche degli RNA cellulari A.F.PalazzoandE.S.Lee,“Non-codingRNA:whatisfunc.onalandwhatisjunk?,”Front.Genet.,vol.6,no.61,p.326,Jan.2015.
I microRNA (miRNA) sono piccoli RNA silenziatori I miRNA sono ncRNA, lunghi 19-24 nt, a singolo filamento.
Ad oggi nell’uomo sono stati identificati 1881 miRNA diversi (www.mirbase.org/)
che si pensa regolino l’espressione di 2/3 dei geni cellulari.
5’
5’
• I miRNA interagiscono con un mRNA bersaglio (target) mediante
appaiamento imperfetto.
• L’espressione dell’mRNA target è inibita.
BIOGENESI dei miRNA miRISC*
siRNA-RISC
miRISC delle piante
*miRNA-Induced Silencing Complex
I geni dei microRNA possono essere unita’ geniche indipendenti o
far parte di geni codificanti per un mRNA Geni miRNA :
• Unita’ geniche indipendenti con un proprio promotore
• Mirtron: Intragenici, situati all’interno delle sequenze introniche di un
altro gene, con cui condividono il promotore I geni dei microRNA possono essere mono o policistronici
I pri-miRNA hanno una struttura comune
Questa struttura ha permesso l’identificazione dei geni dei miRNA
all’interno del genoma
…..cosi’ come i pre-miRNA
I miRNA sono inibitori dell’espressione genica
Ipotetici meccanismi di repressione genica mediati dai miRNA
Krol et al. The widespread regulation of microRNA biogenesis, function and
decay. Nature Reviews Genetics (2010) vol. 11 (9) pp. 597-610
Analisi bioinformatica dei potenziali target di un miRNA
Esistono algoritmi che permettono di predire, per un dato miRNA, gli mRNA
potenziali target, e/o per un dato mRNA i miRNA che potenzialmente lo
possono legare. La predizione si basa sulla complementarieta’ di sequenza,
sulla conservazione evolutiva del complesso, sulla posizione sulla 3’UTR, ecc. Tra gli algoritmi piu’ noti: microRNA.org, targetScan, Pictar, Diana, ecc ecc
Predicted miRNA target sites within the human EDN1 3′UTR (NM_001955) were
determined using microRNA.org Bartel,DavidP."MicroRNAs:genomics,biogenesis,mechanism,andfunc.on.”Cell116.2(2004):281-297.
Un miRNA puo’ controllare l’espressione di centinaia di mRNA
target Silenziamento genico mediato dai miRNA nei mammiferi e nelle piante
mammiferi
piante
La scoperta dei microRNA
I miRNA rappresentano la punta di un iceberg del mondo dei ncRNA e ad oggi sono i
ncRNA meglio caratterizzati
V.Ambros
T.Tuschl
probe compared with the Ddel probe, indicating that lin-4S starts 5 nt upstream of the end
of the Ddel probe.
(B) S1 analysis of total RNA from wild-type N2
using 3! end–labeled rfMGH8 as a probe. The
temperature at which the S1 digestion was performed is indicated above each lane. S1 digestion was for 1 hr. The size of 5! end-labeled
oligonucleotide markers is indicated to the
right.
Lin-4 è il primo miRNA identificato
(Victor Ambros 1993) Cell, Vol. 75, 843–854, December 3, 1993, Copyright ©1993 by Cell Press
The C. elegans Heterochronic Gene lin-4
Encodes Small RNAs
The C. elegans Heterochronic Gene lin-4
Encodes
Small RNAs Complementarity to lin-14
with Antisense
Cell, Vol. 75, 843–854, December 3, 1993, Copyright ©1993 by Cell Press
with Antisense Complementarity to lin-14
Caenorhabditis elegans
Rosalind C. Lee,*† Rhonda L. Feinbaum,* ‡
Ambros and Horvitz, 1987). Animals carrying a lin-4 lossRosalind C. Lee,*† Rhonda L. Feinbaum,* ‡
Ambros and Horvitz, 1987). Animals carrying a lin-4 loss†
†
and
Victor
Ambros
and
Victor
Ambros
tion
or rearrangement
that removes at least
5 kb in(lf)the
lin-4 lin-4(e912),
of-function
(lf) mutation,
lin-4(e912), display reiterations o
of-function
mutation,
display reiterations
of
Harvard
University
early
fates
at
inappropriately
late
developmental
stages;
Harvard
University
region,
including the entire lin-4S- and lin-4L-transcribed
early fates at inappropriately late developmental stages
Department
of Cellular
and Developmental
Biology,
cell lineage patterns normally specific for the L1 are reiterLavoro
citato
8774
volte!
sequences.
To 02138
test
further
the functional
significance
of
Department
of Cellular
and Developmental
Biology,
Cambridge,
Massachusetts
cell execute
lineage
patterns
normally specific for the L1 are reiterated at later stages, and the animals
extra
larval
transcribed sequences,
a noncomplem the lin-4 Massachusetts
Cambridge,
02138we usedmolts
(Chalfie et al., 1981). Theated
consequences
of
these
at later stages, and the animals execute extra larva
heterochronic
developmental
patterns include the abentation screen (see Experimental Procedures)
to isolate
a
et al., 1981). The consequences of these
sence of adult structures (suchmolts
as adult (Chalfie
cuticle and the
novel lin-4 mutation and then identified the
corresponding
vulva) and the prevention of eggheterochronic
laying.
developmental patterns include the abSummary
molecular lesion. Over 20,000 mutagenized
lin-14 nullchromo(0) mutations cause a phenotype opposite to
oflin-4(lf),
adult
structures (such as adult cuticle and the
that of lin-4(lf)
are completelysence
epistatic to
which
somes for
were
screened,
and
a single
novel
lin-4and
allele,
lin-4 is essential
the normal
temporal
control
of
isanimals
consistentwas
with amlin-4 acting as
a negative
regulator
of
diverse ma161,
postembryonic
developmental
events
in ma161
C.
vulva)
and
the prevention
of egg laying.
was identified.
DNA
from
Summary
lin-14 (Ambros and Horvitz, 1987; Ambros, 1989). lin-14(0)
elegans.
lin-4 acts by negatively regulating the level of
plified by PCR and sequenced. The onlymutants
sequence
lin-14
nulland
(0)premutations cause a phenotype opposite to
skip the alterexpression of L1-specific
events
LIN-14 protein, creating a temporal decrease in LIN-14
ation inin the
693larval
bp lin-4
ma161cociously
DNA was
a Cprograms
to
execute
normally
specific
for
the
L2,
protein starting
the first
stage region
(L1). We of
have
that of lin-4(lf) and are completely epistatic to lin-4(lf), which
lin-4 is C.essential
for
theby normal
temporal
control
of
L3, L4,
adult point
stages. lin-14 gain-of-function (gf) mutacloned the
elegans lin-4
locus pair
chromosomal
T transition
at base
517 (see Figure
3).andThis
is high
consistent
with
walking
and transformation
rescue. We
used the C.
diverse
postembryonic
developmental
events
in C.
tions,
which cause
inappropriately
lin-14 activity
at lin-4 acting as a negative regulator o
would
alterother
nucleotide
5
in
lin-4L
and
elegans mutation
clone to isolate
thepresumably
gene from three
late stages
development,
result
in a retarded
phenotype
lin-14
(Ambros
and Horvitz, 1987; Ambros, 1989). lin-14(0
elegans.
lin-4
acts by negatively regulating
the of
level
of
Caenorhabditis
species; all four Caenorhabditis clones
lin-4S.
virtually identical to that of lin-4(lf) (Ambros and Horvitz,
mutants skip the expression of L1-specific events and pre
LIN-14 protein,
a temporal
in LIN-14
functionally
rescue the creating
lin-4 null allele
of C. elegans.decrease
1987). These
observations indicate that in wild-type develComparison of the lin-4 genomic sequence from these
high have
level of lin-14 activity
in the earlyexecute
L1 stage programs normally specific for the L2
cociously
protein starting in the first larval stage opment
(L1). aWe
four species and site-directed mutagenesis of potenspecifies L1-specific programs, and lower levels of lin-14
Transcripts
Are that
Complementary
the
L3, L4, and adult stages. lin-14 gain-of-function (gf) mutacloned
the
C. elegans
lin-4
locus
chromosomal
tial
openlin-4
reading
frames
indicated
lin-4 does
not bytoactivity
in the late L1 specify later stage-specific programs.
encode
a
protein.
Two
small
lin-4
transcripts
of
approx3!UTR
of lin-14
mRNA
walking
and
transformation
rescue. We
used
thedevelopmental
C.
Thus,
the normal
progression
from thecause
exetions, which
inappropriately high lin-14 activity a
imately 22 and 61 nt were identified in C. elegans and
cution
of
L1
programs
to
later
programs
depends
critically
Figure
8.
lin-4
Transcripts
and Complementarity between lin-4 and
The
lin-4
transcribed
sequence
was
combined
in
tandem
elegans
clone
to
isolate
the
gene
from
three
other
found to contain sequences complementary to a relate
stages
of development,
result
in a retarded
phenotype
on
the
lin-4-dependent
decrease
in
lin-14
activity.
LIN-14
è
un
gene
eterocronico,
che
controlla
quando
ed
in
che
successione
devono
avvenire
eventi
lin-14
to the sequence
of the
lin-14
3!UTR
(Wightman et al.,
1991),
peated
sequence
element
in the
3! untranslated
region
Caenorhabditis
species;
all four
Caenorhabditis
clones
virtually
identical
The temporal decrease in lin-14
activity reflects
a de- to that of lin-4(lf) (Ambros and Horvitz
(UTR) ofand
lin-14this
mRNA,
suggesting
thatsearched
lin-4 regulates
(A) Sequences fortiming
lin-4L and
particolari
durante
ilwas
differenziamento
della
larva
(developmental
).lin-4S RNAs, and a proposed secondary
sequence
for
the crease
formation
of lin-4:
in
the
level
of
LIN-14
protein.
LIN-14
protein
is
norfunctionally
rescue
the
lin-4
null
allele
of
C.
elegans.
lin-14 translation via an antisense RNA-RNA inter1987).
These
observations
indicate
that
in wild-type
develstructure
for
lin-4L,and
predicted
the
MULFOLD
program
(see Experilin-14
hybrid RNA
structures,
using the
STEMLOOP
mally
abundant
inprothe nuclei of
late-stage
embryos
Lin-4
è essenziale
un
normale
controllo
temporale
dello
sviluppo
larvalebydi
C.elegans.
action.
Comparison
of the per
lin-4
genomic
sequence
from
these
mental
Procedures).
structure
forin
lin-4S
not shown,
opment
a high
of lin-14
activity
the isearly
L1 stage
younger L1
larvae and then is barely
detectable
by the level
L2A secondary
gram
of the GCGèsequence
analysis package
(Devereux
La
sua
espressione
inversamente
proporzionale
all’espressione
della
proteina
di
to the
of the precise
3! LIN-14
and 5! nucleotides of lin-4S,
four
species
and site-directed
mutagenesis
(Ruvkunof
andpotenGiusto, 1989). lin-14owing
transcripts
areuncertainty
constant
specifies L1-specific programs, and lower levels of lin-14
et al., 1984), as described in Experimental Procedures.
which
affect structural predictions (see text). Sequences
development,
thatsignificantly
lin-14 is negaIntroduction
tial open reading frames indicated that throughout
lin-4 does
not indicating
activity in the late L1 specify later stage-specific programs
Two short blocks of lin-4 sequence were identified (Figure
–86 of IRK1) have
ol (1:1) (American
as dried under N2
orm pure 3H-PIP2
bated with 3H-PIP2
beads. After 1 wash
buffer and counted
. The bound 3H
tal added. For coatidylcholine (PC)
heim) and 90 mg
ied down together
usion proteins were
nd PIP2 antibodies
a further 30 min.
ated by 10% SDS–
ECL (Amersham).
esults. The relative
y serial dilutions of
interactions with Gsa. Biochemistry 28, 611–616 (1989).
Acknowledgements. We thank E. Phan for technical assistance; I. Bezprozvanny, C. Dessauer, D. Logothetis, C.-C. Lu, O. Moe, S. Muallem and H. Yin for discussions and advice; L. Jan for GIRK1 and ROMK1
antibodies; C. Dessauer and A. Gilman for Gai1; P. Casey for Gbg; and R. Alpern for support and
encouragement. This work was supported by grants from the NKF of Texas (C.L.H.) and from the AHA
and NIH (D.W.H.).
activity of other highly related myosin heavy-chain genes . The
unc54C segment has been unique in our overall experience to date:
effects of 18 other dsRNA segments (Table 1; and our unpublished
observations) have all been limited to those expected from previously characterized null mutants.
The pronounced phenotypes seen following dsRNA injection
indicate that interference effects are occurring in a high fraction of
fluorescent generally exp
The mosaic pattern o
was nonrandom. At low
ference in the embryon
when the animal hatch
entiated cells persisted
duced little or no additi
14 postembryonically de
larval stages and these
cells have come throu
versus 8–9 divisions fo
trations of gfp dsRNA,
body-wall muscles, with
cells born during both em
The non-striated vulval
development, appeared
concentrations of inject
We do not yet know
ference in C. elegans. Som
about possible targets an
First, dsRNA segmen
promoter sequences d
(Table 1). Although con
tional level, these exper
level of the gene.
Second, we found th
nounced decrease or eli
script (Fig. 3). For this ex
3) that is abundant in
straightforward in situ h
genous mex-3 mRNA
dsRNA segment derive
which purified mex-3 a
stantial endogenous mR
Third, dsRNA-media
to cross cellular bounda
lacZ) into the body cavit
robust interference wit
(Table 2). Interference
arms, ruling out the occ
1998: Fire and Mello identificano l’RNA interference in Caenorhabditis elegans
Correspondence and requests for materials should be addressed to C.L.H. (e-mail: chuan1@mednet.
swmed.edu).
Potent and specific
genetic interference by
double-stranded RNA in
Caenorhabditis elegans
a
b
c
d
Andrew Fire*, SiQun Xu*, Mary K. Montgomery*,
Steven A. Kostas*†, Samuel E. Driver‡ & Craig C. Mello‡
* Carnegie Institution of Washington, Department of Embryology,
115 West University Parkway, Baltimore, Maryland 21210, USA
† Biology Graduate Program, Johns Hopkins University,
3400 North Charles Street, Baltimore, Maryland 21218, USA
‡ Program in Molecular Medicine, Department of Cell Biology,
University of Massachusetts Cancer Center, Two Biotech Suite 213,
373 Plantation Street, Worcester, Massachusetts 01605, USA
Figure 3 Effects of mex-3 RNA interference on levels of the endogenous mRNA.
2001: il lab di T.Tuschl identifica in diversi organismi (uomo, topo, Drosofila)
diversi miRNA simili, in struttura e funzione, a lin-4 di C. elegans
+
n of ROMK1 K channel
077–8081 (1994).
ir2.1 inward rectifier K+
s. Neuron 13, 1413–1420
tructure and functional
re 364, 802–806 (1993).
nd molecular properties.
multimer of two inwardly
+
y rectifying K channels.
nkage of the cardiac ATP96).
TP potassium channels by
hibits oncogene-induced
al expression of a mouse
ated potassium channel.
channel by a G-protein-
coupled to GTP-binding
ficient in Lowe syndrome
Sci. USA 92, 4853–4856
Interference contrast micrographs show in situ hybridization in embryos. The
1,262-nt mex-3 cDNA clone20 was divided into two segments, mex-3A and mex-
3B, with a short (325-nt) overlap (similar results were obtained in experiments with
no overlap between interfering and probe segments). mex-3B antisense or
.........................................................................................................................
dsRNA was injected into the gonads of adult animals, which were fed for 24 h
Experimental introduction of RNA into cells can be used in
certain biological systems to interfere with the function of an
endogenous gene1,2. Such effects have been proposed to result
from a simple antisense mechanism that depends on hybridization between the injected RNA and endogenous messenger RNA
transcripts. RNA interference has been used in the nematode
Caenorhabditis elegans to manipulate gene expression3,4. Here we
investigate the requirements for structure and delivery of the
interfering RNA. To our surprise, we found that double-stranded
RNA was substantially more effective at producing interference
than was either strand individually. After injection into adult
animals, purified single strands had at most a modest effect,
whereas double-stranded mixtures caused potent and specific
interference. The effects of this interference were evident in
both the injected animals and their progeny. Only a few molecules
of injected double-stranded RNA were required per affected cell,
arguing against stochiometric interference with endogenous
before fixation and in situ hybridization (ref. 5; B. Harfe and A.F., unpublished
Nature © Macmillan Publishers Ltd 1998
NATURE | VOL 391 | 19 FEBRUARY 1998
observations). The mex-3B dsRNA produced 100% embryonic arrest, whereas
.90% of embryos produced after the antisense injections hatched. Antisense
probes for the mex-3A portion of mex-3 were used to assay distribution of the
endogenous mex-3 mRNA (dark stain). four-cell-stage embryos are shown;
similar results were observed from the one to eight cell stage and in the germ
line of injected adults. a, Negative control showing lack of staining in the absence
of the hybridization probe. b, Embryo from uninjected parent (showing normal
pattern of endogenous mex-3 RNA20). c, Embryo from a parent injected with
purified mex-3B antisense RNA. These embryos (and the parent animals) retain
the mex-3 mRNA, although levels may be somewhat less than wild type. d,
Embryo from a parent injected with dsRNA corresponding to mex-3B; no mex-3
RNA is detected. Each embryo is approximately 50 mm in length.
Table 2 Effect of site of injection on interference in injected animals and their progeny
dsRNA
Site of injection
Injected-animal phenotype
Gonad or body cavity
Gonad or body cavity
No twitching
Strong nuclear and mitochondrial GFP expression
Gonad
Body-cavity head
Body-cavity tail
Weak twitchers
Weak twitchers
Weak twitchers
gfpG
gfpG
Gonad
Body-cavity tail
Lower nuclear and mitochondiral GFP expression
Lower nuclear and mitochondrial GFP expression
lacZL
lacZL
Gonad
Body-cavity tail
Lower nuclear GFP expression
Lower nuclear GFP expresison
...................................................................................................................................................................................................................................................
None
None
unc22B
unc22B
unc22B
Rare
Rare
...................................................................................................................................................................................................................................................
Esiste una correlazione diretta tra il numero dei miRNA espressi da una
data specie e la sua complessita’ morfologica
Kosik.MicroRNAstellanevo–devostory.NatureRevNeurosc(2009)10:pp.1-6
miRNA e Tumori
Alterazioni nell’espressione di specifici miRNA sono responsabili dell’inizio e
della progressione di numerosi tumori umani.
miR-15 e miR-16 sono sono soppressori tumorali; miR-17-92 sono onco-miRNA
REVIEWS
Epigenetic regulation of miR
hypomethylation, CpG island
histone-modification losses repr
of malignant transformation76. T
Delezione
osservata
nella
have investigated
whether
such e
leucemia
linfociti
cronica
expression. Scott et al. showed t
umana
(CLL)
histone
deacetylase inhibition i
sive and rapid alteration of miRN
Saito et al. found that the combi
bladder cancer cells with 5-aza-2
CdR) and the histone deacety
4-phenylbutyric acid
(PBA) has
Amplificazione
osservata
expression of miRNAs78. Sevent
nei linfomi umani, ad es
screened by a microarray assay)
B-cell diffuse large cell
than threefold, and miR-127 wa
lymphomas
(DLBLs)
expressed. This miRNA is loc
chromosome 14q32, a region th
types of translocations identified
cers and deleted by LOH in solid
the combined treatment was acc
Figure 3 | Chromosomal alterations at microRNA loci. The main chromosomal
in DNA methylation and an in
alterations
microRNA
loci, loss of
heterozygosity
and (November
amplification,2006)
are | doi:10.1038/nrc1997
Calinatand
Croce (miRNA)
Nature Reviews
Cancer
6, 857–866
markers around the transcripti
identified at two separate regions of chromosome 13. a | Shows the 13q14.3 deletion
miRNA nel sistema nervoso
•
•
•
•
•
•
circa il 70% dei miRNA è espresso nel cervello e molti miRNA sono
neuro-specifici
nel sistema nervoso i livelli di espressione dei miRNA è maggiore
che in altri tessuti
i miRNA sono coinvolti nello sviluppo del sistema nervoso e nella
morfogenesi dei neuroni (nella crescita dei neuriti e nella
formazione delle spine dendritiche)
contribuisono al controllo delle funzioni sinaptiche e della
plasticita’ nell’adulto
la loro de-regolazione è stata osservata in quasi tutte le malattie
neurologiche studiate
sono considerati potenziali biomarker e target terapeutici nei
disturbi neurologici
miRNA alle sinapsi
Schrad,Gerhard."microRNAsatthesynapse."NatureReviewsNeuroscience10.12(2009):842-849.
….caratteristiche degli RNA cellulari A.F.PalazzoandE.S.Lee,“Non-codingRNA:whatisfunc.onalandwhatisjunk?,”Front.Genet.,vol.6,no.61,p.326,Jan.2015.
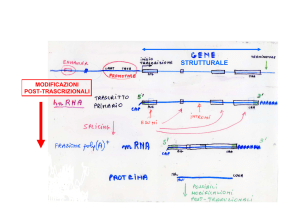
I LONG NON CODING RNA (lncRNA)
Caratteristiche
Funzioni biologiche dei lncRNA (ipotetiche)
A.
B.
C.
D.
E.
F.
G.
sequestrare fattori di trascrizione
sequestrare i microRNA (miRNA sponges)
possono essere componenti di complessi RNA-Proteine (RNP) reclutare rimodellatori e modificatori della cromatina, come nel caso di Xist
modulare lo splicing inibire la traduzione bloccando l’mRNA
indurre la degradazione dell’ mRNA Funzioni biologiche dei lncRNA (dimostrate)
ncRNA ed evoluzione del SNC
ScienCstsIdenCfyGeneDifferenceBetween
HumansandChimps
Si ipotizza che i ncRNA abbiamo un ruolo chiave nella rapida evoluzione del SNC umano. In uno studio del 2006 sono state identificate e analizzate le human accelerated regions
(HARs), regioni del genoma che mostrano un accumulo di mutazioni significativamente
accellerato nel periodo evolutivo che corrisponde all’evoluzione del nostro antenato dallo
scimpanze’. Molte di queste HAR trovate sono associate con geni coinvolti nella
regolazione della trascrizione e dello sviluppo del sistema nervoso. HAR1, l’elemento variato piu’ significativamente, è parte di un gene codificante per
ncRNA, localizzato sul cromosoma 21 ed espresso durante lo sviluppo corticale dell’uomo.
PollardKS,SalamaSR,LambertN,LambotM-A,CoppensS,PedersenJS,etal.AnRNAgeneexpressedduringcor.caldevelopmentevolved
rapidlyinhumans.Nature.2006Aug16;443(7108):167–72.
Pon.ngCP,LunterG.Evolu.onarybiology:humanbraingenewinsgenomerace.Nature.2006Sep14;443(7108):149–50.
Tes.
BiologiaMolecolare,Amaldietal
BiologiaMolecolaredelgene,Watsonetal
Reviews
Bartel,DavidP."MicroRNAs:genomics,biogenesis,mechanism,andfunc.on.”
Cell116.2(2004):281-297.
CalinandCroceNatureReviewsCancer6,857–866(November2006)|doi:10.1038/
nrc1997
Schrad,Gerhard."microRNAsatthesynapse."NatureReviewsNeuroscience10.12
(2009):842-849.