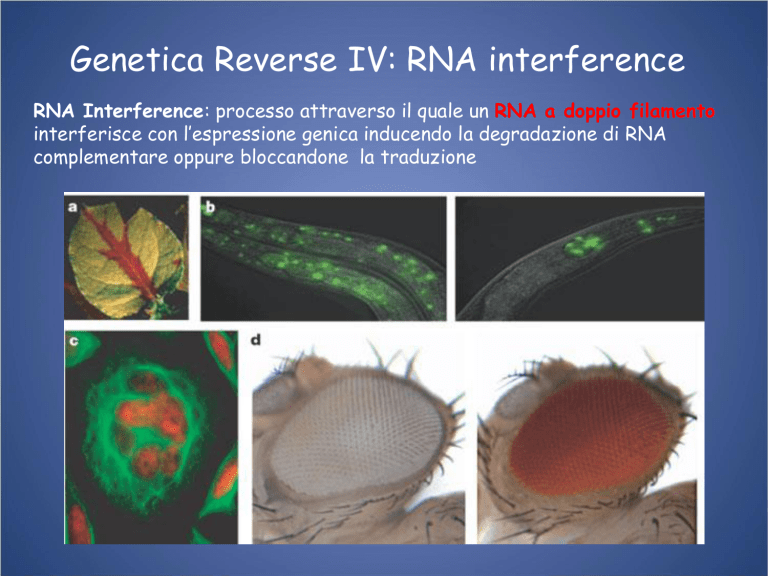
Genetica Reverse IV: RNA interference
RNA Interference: processo attraverso il quale un RNA a doppio filamento
interferisce con l’espressione genica inducendo la degradazione di RNA
complementare oppure bloccandone la traduzione
COSUPPRESSION IN PETUNIA: overexpression of chalcone synthase
CHS gene (anthrocyanin pigment gene) in petunia caused the loss of
pigment in flower sectors. (Napoli et al.1990)
The levels of endogenous
as well as introduced CHS
were 50-fold lower than
in wild-type petunias,
which led the authors to
hypothesize that the
introduced transgene was
“cosuppressing” the
endogenous CHS gene.
dsRNA
- occurrence of aberrant transcripts due to overexpression
- formation of dsRNA from aberrant transcripts by RdRP (RDR6)
-formation of siRNAs that silence both transgene and internal gene
1992: Quelling in Neurospora crassa.
Cogoni and Macino introduced a gene (al1) needed for carotenoid synthesis
in Neurospora crassa:
The introduced gene led to inactivation of the mold's own gene in about
30% of the transformed cells. They called this gene inactivation "quelling."
Model for post‐transcriptional gene
silencing in Neurospora. dsRNA can
be produced either directly from
inverted repeats or indirectly
whereby an RdRP (QDE1) makes a
complementary strand of an
aberrant single‐stranded (ss)RNA
produced from the transgene. The
aberrant RNA is a result of an
epigenetic modification of the
transgenic locus driven by the
repetitive nature of the transgene
and mediated by QDE3. Before
Dicer processes the dsRNA into the
siRNAs, the dsRNA is used as a
template by RdRP (QDE1) to
synthesize a diffusible factor. The
siRNAs are subsequently
transferred by QDE2 from Dicer to
the mRNA‐degrading enzyme. See
text for additional details.
Paramutazione
(Brink, 1968; Coe and Hagemann 1968)
La paramutazione è un cambiamento epigenetico
ereditabile nello stato di espressione di un allele dovuto
all’interazione con il suo allele omologo.
Trans-interazione tra 2 sequenze omolghe che causa
una modificazione epigenetica ereditabile nello stato si
espressione genica
Mais: paramutazione al locus b1 (booster 1)
Alleli coinvolti nella paramutazione:
B’ (allele paramutagenico) è poco espresso
B-I (allele paramutabile) è altamente
trascritto ed è responsabile della
pigmentazione viola della pianta
MOP1
la capacità paramutagenica di B’ è
estremamente stabile ed ha una
penetranza del 100%. Gli epialleli B-I e
B’ sono strutturalmente identici nella
sequenza nucleotidica ma mostrano un
pattern di metilazione differenziale e
una diversa sensibilità alla digestione
con nucleasi in corrispondenza di una
regione mappata per ricombinazione 100
kb a monte del sito di inizio della
trascrizione costituita da una sequenza
di 853 paia di basi ripetuta in tandem
sette volte.
Il diverso stato epigenetico di queste
sequenze ripetute nei due tipi di
epialleli è in grado sia di garantire sia
la capacità paramutagenica di B’ che
l’elevato tasso di trascrizione di B-I
Forward genetics provides answers!
Screen for mutants that maintain purple pigment in the
presence of B’
modifier of paramutation mop1-1 mutation
(A) B' Mop1+/mop1-1
(B) B' mop1-1/mop1-1
should look like A; instead looks like C
(C) B-I Mop1+/Mop1+
Positional cloning of the affected gene - mop1 encodes an RNA dependent
RNA polymerase
What mechanism of paramutation is suggested by this finding?
(Dorweiler et al. Plant Cell 12:2101; Alleman et al. Nature 42:295)
Paramutation at the maize b1 locus
The two alleles that participate in
paramutation at the b1 locus are identical in
sequence and contain an identical control
region consisting of seven tandem repeats
(red and white boxes). However, the B-I
allele is highly transcribed while the B′ allele
is not. The two alleles exhibit epigenetic
differences in chromatin structure, histone
modifications, and DNA methylation and may
be associated with distinct proteins that
maintain these epigenetic states. The
tandem repeats are bidirectionally
transcribed in both B-I and B′ plants,
producing repeat RNA that then forms
dsRNA and is processed into siRNAs. The
proteins MOP1, RMR6, and MOP2 are
important for the production and
amplification of the dsRNA and siRNAs. The
siRNAs are hypothesized to direct chromatin
modifications at the tandem repeats via
mechanisms and proteins that are currently
unknown, but this process is blocked at the
B-I allele, potentially by the active chromatin
state, bound proteins, or nuclear
environment. Paramutation occurs in
heterozygous plants, when the highly
transcribed B-I allele is “paramutated”, or
converted, to the silenced B′ state. siRNAs
produced from the tandem repeats are
hypothesized to mediate trans-interactions or
communication between the alleles, as well
as direct the establishment of a closed
chromatin structure at the B-I tandem
repeats. The conversion of B-I to a silenced
epigenetic state is meiotically stable, and in
the next generation all progeny will inherit a
silenced B′ allele. The newly paramutated
allele is termed B′*.
Genetics Research International
Volume 2012 (2012), Article ID 689819,
1998: Potent and specific genetic interference by double-stranded RNA in
Caenorhabditis elegans
ANDREW FIRE*, SIQUN XU*, MARY K. MONTGOMERY*,
STEVEN A. KOSTAS*†, SAMUEL E. DRIVER‡ & CRAIG C. MELLO‡
Unc-22 (Uncoordinated 22)
•Codes for a non essential myofilament
•It is present several thousand copies/cell
Injection for RNAi
• 6-10 adult hermaphrodites were
injected with 0.5x106-1x106 molecules
into each gonadal arm.
Unc-22 phenotype
• 4-6 hours after injection,
eggs collected.
• Screened for phenotypic
changes
– twiching
Exon
Size
RNA
Phenotype
Exon 21-22
742
Sense
Antisense
Sense+antisense
Wildtype
Wildtype
Twicher (100%)
Exon 27
1033
Sense
Antisense
Sense+antisense
Wildtype
Wildtype
Twicher (100%)
Mex-3
• mex-3 encodes two RNA binding proteins; in the early
embryo, maternally provided
• Mex-3 is required for specifying the identities of the
anterior AB blastomere and its descendants, as well as
for the identity of the P3 blastomere and proper
segregation of the germline P granules
Mex-3 RNAi
b, Embryo from uninjected
parent (showing normal pattern
of endogenous mex-3 RNA20).
c, Embryo from a parent injected
with purified mex-3B antisense
RNA. Retain the mex-3 mRNA,
although levels may be somewhat
less than wild type.
d, Embryo from a parent injected
with dsRNA corresponding to
mex-3B; no mex-3 RNA is
detected.
RNA interference (RNAi)
gene silencing at
•transcriptional level (TGS)
(transcriptional gene silencing)
methylation of promoter, heterochromatin formation
- preventing interaction of transcription factors
• posttranscriptional level (PTGS)
(posttranscriptional gene silencing)
- transcript cleavage
- block of translation
General mechanisms of RNA interference
Small silencing RNAs
Biogenesi dei microRNA
Characteristics of Drosophila piRNAs
(A) Radioactively labeled RNA isolated under identical
conditions from specific Piwi-family RNPs and Ago1
was analyzed on a denaturing polyacrylamide gel. The
positions of RNA size markers, electrophoresed in
parallel, are shown to the left. Indicated are piRNAs
(solid arrowhead), miRNAs (open arrow head), and 2S
rRNA (arrow), which is also present in purifications
using control antibodies.
(B) Size distributions of sequenced piRNAs specifically
bound by the three Piwi-family members.
(C) Pie chart summarizing the annotation of piRNA
populations in total RNA and those bound by Piwi, Aub
and Ago3
Figure 2 Characteristics of Drosophila piRNAs (A) Radioactively labeled RNA isolated under identical conditions from specific Piwifamily RNPs and Ago1 was analyzed on a denaturing polyacrylamide gel. The positions of RNA size markers, electrophoresed i...
Julius Brennecke , Alexei A. Aravin , Alexander Stark , Monica Dus , Manolis Kellis , Ravi Sachidanandam , Gregory...
Cell, Volume 128, Issue 6, 2007, 1089 - 1103
http://dx.doi.org/10.1016/j.cell.2007.01.043
piRNA biogenesis in Drosophila and mice consists of the primary piRNA processing pathway
and the amplification loop.
Ishizu H et al. Genes Dev. 2012;26:2361-2373
Copyright © 2012 by Cold Spring Harbor Laboratory Press
Biogenesis of piRNA
RNAi and DNA
methylation in
Arabidopsis
RNAi and Heterochromatin formation
in S.pombe
RNA interference in Drosophila
Microiniezione di dsRNA
in embrioni
Mutante twist
Mutante engrailed
RNAi twist
RNAi engrailed
..oppure
Espressione in vivo di
sequenze ripetute
invertite omologhe al
gene da silenziare
Misquitta e Paterson (1999)
Genome-wide RNAi in Drosophila
Librerie di dsRNA disponibili in Drosophila
Nature. 2007. 448(7150):151-6
A genome-wide transgenic RNAi library
for conditional gene inactivation in Drosophila
Georg Dietzl1,2, Doris Chen1, Frank Schnorrer2, Kuan-Chung Su1, Yulia Barinova1, Michaela
Fellner1,2, Beate Gasser1, Kaolin Kinsey1,2, Silvia Oppel1,2, Susanne Scheiblauer1, Africa
Couto2, Vincent Marra1, Krystyna Keleman1,2 & Barry J. Dickson1,2
1.Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Dr. Bohr-Gasse 3-5,
A-1030 Vienna, Austria
2.Research Institute of Molecular Pathology (IMP) Dr. Bohr-Gasse 7, A-1030 Vienna, Austria
Espressione in vivo di sequenze ripetute invertite
omologhe al gene da silenziare
Construction of a genome-wide transgenic RNAi library
Moscerini transgenici vengono trasformati con costrutti opportunemente
ingegnerizzati per esprimere, sotto il controllo del promotore eterologo di
lievito, frammenti corrispondenti al gene da silenziare clonati come repeats
invertite
pMF3 contiene:
10 GAL4-responsive elementi UAS
Il promotore basale di hsp70
150 bp del secondo introne di ftz
Il segnale dipoliadenilazione di SV40
pMF3
Construction of a genome-wide transgenic RNAi library
Coppie di primers sono stati opportunamente disegnate per amplificare mediante PCR
porzioni diverse di ogni “predicted protein-coding gene” nella sequenza genomica di
Drosophila.
• 77.8% dei costrutti rappresentano in maniera specifica un unico esone codificante
• 22.2% dei costrutti rappresentano esono diversi dello stesso gene oppure 5’ o 3’ UTR
•In totale, I 15,072 costrutti UAS-IR, rappresentano 13,327 geni differenti
le dimensioni delle repeat invertite variano da109 a 415 base pairs (bp), con una media
di 323 bp.
Predizione dei potenziali target di ciascun costrutto RNAi
vengono definite tutte le possibili combinazioni di siRNA (da 19bp)
potenzialmente prodotte da ogni RNA hairpin
Se tutti I 19-mers prodotti dal costrutto UAS-IR corrispondono in maniera
univoca al gene target, si ha un unico ON TARGET e nessun OFF TARGET.
dei circa 15000 costrutti il 96.9% ha un singolo ON TARGET.
I costrutti sono stati usati per trasformare la linea germinale di ceppi di
moscerini
Ciascuna inversione è stata verificata per PCR e mappata
in totale sono state ottenute 22,270 linee transgeniche, rappresentative di
13,251 RNAi costrutti e di 12,088 genes (88.0% dei geni di Drosophila).
efficienza di RNA knockdown
•64 linee linee UAS-IR
In 25 delle 64 linee analizzate il
messaggero del gene target è
ridotto di più del 50%
Analisi dei fenotipi
Gene activity does not become null:
Hypomorphic effect
Transformer 2 (tra2) RNAi and
mutant females anatomically
resemble males, including male
genitalia and abdominal
pigmentation.
The eyes are greatly reduced or
absent in eyes absent (eya) RNAi
and mutant males.
Stubble (Sb) males have short,
stubby bristles on the notum.
Dicer-2 aumenta l’efficienza di RNAi
Conclusions
Proof-of-principle for genome-wide tissue-specific RNAi screens in
Drosophila
RNAi transgenes consist of short gene fragments cloned as inverted
repeats and expressed using the binary GAL4/UAS system.
22,270 transgenic lines were generated, covering 88% of the
predicted protein-coding genes in the Drosophila genome.
Over 60% of the transgenic lines trigger potent and specific gene
interference.
Up to 90% may be functional in combination with the appropriate
drivers, assays, and RNAi-enhancing tools such as UAS-Dcr-2.