PhD-IBMG-II-Matinée
Genetic information
Genomi, geni, organizzazione comparativa
Funzioni, famiglie, mutazioni, evoluzione
Mutazione e riparazione del DNA
NGS
Alternative splicing
Il genoma umano 3,200 Mb
23 cromosomi (x2)
The Human Genome Project
Animated tutorials on the Human
Genome Project:
http://www.genome.gov/Pages/
EducationKit/
(free downloads or on-line view)
1. DNA sequence
Sito NCBI Genomes Eukaryotic (Mammals)
Genomic Data
1990-2003
Human Genome Project
2001
The HGP consortium and Celera release the first draft of
95% Human Genome
2003
Sequencing is completed
2001-today
Several other genomes sequenced
NGS technology
2007
The ENCODE project releases results on 1% human genome
2012
The ENCODE project publishes complete results
Table 1-1 (part 2 of 2) Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
prokaryotes
5x
50x
Eukaryotes and Prokaryotes
different genome organization
different gene structure
The rationale for genom organization in Prokaryotes and Eukaryotes is
fundamentally divergent
Prokaryotes: essentials, reproductive speed, high mutation rate
Eukaryotes: diversification, adaptation, reassortment
Let’s see first some common traits:
The overall «order» of transcription units (see below for definition) does
not follow a simple logic.
Transcription Units are in both orientations on the chromosomes
Noncoding parts are present that are either functional to chromosome
dynamics (e.g. origins of replication) or mobile genetic elements (e.g.
virus, transposons).
Transcription Units are in both orientations on the chromosome
Transcription Unit (T.U. or TU) is a part of DNA that is found transcribed
in RNA in at least some circumstances. A «promoter» or «promoter-like»
sequence always flanks 5’ a TU.
5’
3’
P
5’
3’
Why do we say «TU» and not «gene».
TU is a physical entity, experimentally proven.
The «gene» is a concept
Definitions of «gene»
-
One gene, one character
One gene, one protein
One gene, some protein isoforms (after alternative splicing)
One gene, one molecular function
One gene one functional module (Molecular Biology)
One gene one Transcriptional Unit (Genomics)
Protein coding genes
Even though proteins that perform similar functions are extraordinarily conserved,
genes may be remarkably different in genomic organization
Essentials:
promoter
ORF
terminator
Prokaryotic genes essentially follow the operon model
leader or 5’UTR
DNA
5’
tail or 3’-UTR
ORF2
ORF1
Transcriptional
termination
ORFn
This part is
copied into RNA
(transcription)
Coding A
Coding B
promoter
(position site for
RNA Polymerase)
Coding N
Spacers (0-few bp)
ATG 1st codon
operator (regulatory element)
Stop codon
Transcribed RNA is «polycistronic» (i.e. contain more than one string of information
Un esempio molto noto in E.coli: l’operone del lattoso
LacZ: -galactosidase cleaves lactose to galactose + glucose
Lac Y: lactose permease, pumps in lactose against electrochemical gradients
LacA: Thiogalactoside transacetylase.
Lac I: lac repressor
Eukaryotes
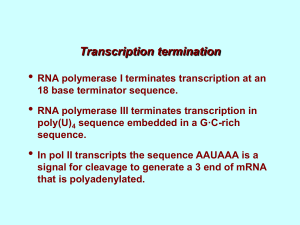
Protein coding genes are often discontinuous.
Coding sequences (exons) are interrupted by noncoding sections (introns)
Transcribed RNAs can be mono-cistronic or polycistronic
3’utr
For large eukaryotic genomes, the presence of introns is a complication.
In this case we should consider separately two distinct entities:
1. The genomic sequence (i.e. the Transcription Unit)
2. The RNA produced after processing of the primary RNA transcript
(mRNA in the case of protein-coding genes)
Eukaryotes: monocystronic
parts remaining in mRNA
5’
Intron N
Intron 1
poly(A) signal
Intron 2
Regulatory
region and
promoter
3’-UTR
(regulatory)
Exon 1
Exon 2
this part is copied in primary RNA
Exon 3
Exon N
Il processo di splicing congiunge le diverse parti che si ritroveranno nell’RNA
messaggero
Comparative:
Human
Yeast
Drosophila
Mais
A 50 Kb tract of the Human genome
25K
50K
H. sapiens
Genoma umano
3200 Mb
DNA intragenico
2000 Mb
Sequenze di geni e genecorrelate 1200 Mb
Coding
Sequenze
correlate
1152 Mb
48 Mb
Pseudogeni
Ripetizioni
intersperse 1400
Mb
Altre regioni
intrageniche
600 Mb
Introni, UTR
Frammenti
genici
Microsatelliti
90 Mb
LINE 640
Mb
Elementi LTR
250 Mb
Varie
510 Mb
SINE 420
Mb
Trasposoni DNA
90 Mb
Interspersed repetitive elements - Mobile genetic elements
pseudogenes
Second class of pseudogenes
are gene copies inactivated
by multiple mutations, or:
Retrotranscription-insertion
Genes
Protein coding (mRNA)
noncoding: ncRNA
Range
1–cent.
0-cent.
Range
30- some Kb
300- some Mb
100- some Kb
-
z
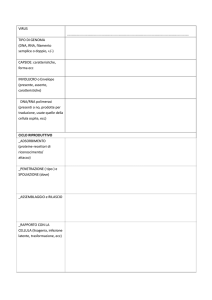
I valori mostrati in tabella sono i valori medi
2Kb- 100Mb
Range (appr.)
While genes vary enormously in size from bacteria to mammals, due to intronic
prevalence, coding regions (ORF) are quite uniform, possibly due to protein
structural constraints.
Predicted ORF products mean size in completely sequenced organisms
Average a.a. 128 Da
in peptides: 110 Da
Functional
catalogue
Gene functions
Protein coding genes are organized in Gene families
Bacillus subtilis
Figure 1-24 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Duplicazioni per ricombinazione diseguale
Traslocazioni
Duplicazioni esoniche
Figure 1-23 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Figure 1-25a Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Figure 1-25c Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008)
Le similitudini tra proteine hanno rivelato un addizionale
livello di organizzazione:
il dominio
Il dominio è una sottostruttura prodotta da qualunque parte
del polipeptide che si possa ripiegare in una conformazione
stabile indipendentemente dal resto della proteina.
Il concetto di dominio è molto importante in genomica,
perchè spesso i domini delle proteine sono codificati da
singoli esoni, giustificando la teoria dell’”exon shuffling” per
l’evoluzione delle proteine.
Mutation & Repair
Vai a 33
M-Phase
Ripasso da Biologia della Cellula
Ripasso da Biologia della Cellula
La domanda che affronteremo oggi è:
Come si fronteggia la possibilità di errore durante la fase di replicazione
del DNA ?
Come viene risolto il problema di eventuali danneggiamenti chimicofisici del DNA in cellule stazionarie ?
Mutazioni e riparazione del DNA
Mantenere inalterato il proprio patrimonio genetico e
passarlo inalterato alla discendenza è un necessità assoluta
per gli organismi viventi.
Qualsiasi irregolarità durante la replicazione del DNA oppure
qualsiasi danno chimico-fisico che succeda al DNA in fase non
replicativa
è potenziale fonte di mutazione
Tutti gli organismi mantengono il più possibile inalterato il DNA,
tramite complicati e dispendiosi meccanismi di riparazione e
manutenzione
cambio di una base
5’
5’
5’
5’
5’
DNA
mutato
scivolamento durante la replicazione
Risultato di una mutazione
In aploidi (es.: E. coli)
se tollerata
si fissa nella discendenza
se non tollerata
la cellula muore
In eucarioti pluricellulari (es. Uomo):
In cellule somatiche
se tollerata
a) nulla
b) la cellula trasforma (cancro)
In cellule germinali
se non tollerata
la cellula muore
se tollerata
si fissa nella discendenza
se non tollerata
a) la cellula muore
b) lo sviluppo non è possibile
Esempi:
Riarrangiamenti o ricombinazioni (grandi mutazioni)
Transposition = change in the order of two or more sequences
Crossing-over = exchange of fragments between DNA molecules
Deletion = loss of a fragment of DNA
Duplication = replication of a fragment of DNA
Amplification = repeated replication of a DNA fragment
Insertion = addition of a fragment of DNA within a sequence
Il “peso” di una mutazione dipende dal contesto in cui la mutazione avviene
All’interno di una sequenza codificante:
Il codice genetico è ridondante
Deletions & insertions
(= 1 a.a.)
(indels)
La proteina perderà un aminoacido,
ma il resto è invariato.
(reading frame changed)
La proteina cambia totalmente a
partire dal punto della mutazione
Frequenza dei cambiamenti
La velocità con la quale si fissano le
mutazioni dipende da quanto la funzione
di ogni particolare tratto di DNA (per es.:
un gene) dipende dalla sua sequenza
nucleotidica.
La fedeltà di mantenimento della sequenza del
proprio DNA è essenziale per ogni forma di vita
Fedeltà nella replicazione
Replicazione senza “proofreading” (3’-5’ esonucleasi): 1 / 104 - 105
Replicazione con proofreading:
1 / 107 – 108
Riparazione degli errori replicativi:
Mismatch repair
1 / 109
Riparazione dei danni accidentali in fase non replicativa
(vari)
Mantiene la frequenza di mutazione a < 1 / 109
1° categoria:
Mutazioni che avvengono in fase replicativa, per errori del replisoma che
non vengano riparati immediatamente dalla DNA Polimerasi stessa, con il
meccanismo di “proofreading” (3’5’ esonucleasi)
Errori dovuti a errata incorporazione: attività proofreading (3’-5’-esonucleasi) della DNA Polimerasi
(vedi: Replicazione)
1° categoria: Errori residui in replicazione (1 / 107 – 108).
Vengono riparati dal meccanismo del Mismatch Repair
(riparo dell’appaiamento scorretto).
E. coli: sistema enzimi MutS, MutL, MutH.
Uomo: enzimi MSH (MutS Homologs), MLH, PMS
Mismatch
T:G
MutS
MutL
MutH
ATP
DNA Pol. I
DNA Ligase
DNA ligasi salda l’ultima interruzione
Distinzione del filamento “vecchio” da
quello neosistetizzato in E. Coli.
E. Coli metila tutte le
sequenze 3’-GATC-5’
(dam metilasi)
Il DNA neosintetizzato per un
po’ resta libero da metilazioni
MutH taglia il filamento
non metilato
Eucarioti: esistono sistemi molto simili.
Uomo: enzimi MSH (MutS Homologs)
la predisposizine genetica al tumore al colon è dovuta a mutazioni in uno dei
geni che codificano proteine del sistema (MSH2)
Il sistema della dam metilasi, tuttavia, è limitata a E. coli. Non si sa ancora con
esattezza come, negli organismi superiori, il sistema riconosca il filamento
neosintetizzato.
2° categoria:
Mutazioni post-replicative
1.
Alterazioni spontanee
2.
Alterazioni dovute a mutageni
Post-replicative DNA repair is active in all cells,
including terminally differentiated cells such as
myocytes or neurons that will never replicate
Idrolisi del legame glicosidico e perdita della base
La rimozione di purine
(max: G) è massiva: in
un giorno, dal genoma
di un mammifero
vengono perse circa
10.000 purine / cellula.
Alterazioni spontanee
deossiriboso
Negli organismi superiori, molte
C sono metilate: una C metilata
se deaminata dà una T.
Per evitare l’accumulo di
mutazioni, esiste un sistema di
riparo che, in presenza di
appaiamenti errati T:G, rimuove
selettivamente le T, riportando
la situazione alla normalità.
Base deamination is spontaneous: circa 100 bases / day / cell.
Some chemical compounds increase the rate of deamination, such as nitrous acid
(deaminates A, C and G) or sodium bisulfite (deaminates C)
Pairs with C
rather than T
Cytosine deamination uracil
(pairs with A rather than G)
Guanine deamination xantine (blocks replication)
radiation
UV
analoghi delle basi
agenti intercalanti
Riparazione
Talune modificazioni possono essere corrette in situ da appositi enzimi
Riparazione dei dimeri di timina con fotoliasi (E.coli) (no eucarioti)
(Base excision repair – BER)
Riparazione mediante escissione di base
La maggior parte delle piccole modificazioni delle basi, come
deaminazioni, alchilazioni etc., sono riconosciute da una batteria di enzimi
appositi che continuamente percorrono il DNA, trovano le alterazioni e
rimuovono le basi danneggiate tagliando il legame glicosidico.
Le DNA glicosilasi
(Base excision repair – BER)
Dopo il taglio, il sito viene
riconosciuto da speciali endonucleasi,
chiamate AP-endonucleases
che rimuovono lo zucchero dallo
scheletro del DNA.
In seguito DNA polimerasi e DNA
ligasi riparano il “gap”.
Alcuni agenti fisici o composti mutageni inducono cambiamenti più
grandi, che danno deformazioni più importanti nella struttura della
doppia elica.
(Alchilazioni, dimeri di timina, intercalanti)
Questi vengono riparati con il meccanismo della
NER (nucleotide excision repair).
NER nucleotide
excision repair
(E.coli)
NER
E.coli: Uvr A, UvrB, UvrC, UvrD
H. sapiens: XPC, XPA, XPD, XPF (ERCCI), XPG
(dove “XP” sta per xeroderma pigmentosum)
Mutazioni puntiformi e
riarrangiamenti
1) durante la fase di replicazione del DNA
1-Proofreading
2-Mismatch Repair
2) Non dipendenti dalla replicazione del DNA
- spontanee
- da agenti esterni
1-Base excision repair
2-Nucleotide excision repair
3-Direct reversal
Se la lesione nel DNA non viene riparata .....
DNA Polimerasi “trans-lesione”
Generazione di “double strand breaks” (DSB)
Se la lesione nel DNA non viene riparata .....
DNA Polimerasi translesione: famiglia Y
In figura, sono rappresentate
le proteine di E. coli
DSB=double strand break
Se la lesione riguarda entrambi i filamenti ?
riparazione di tipo ricombinativo
Ricombinazione Omologa
Concetto di omologia – concetto di identità
Ricombinazione generale o ricombinazione omologa
In caso di danni del doppio filamento (DSB)
?
Il meccanismo di riparazione funziona
schematicamente come se ognuno dei due
filamenti lesi andasse a recuperare lo
“stampo” dal cromosoma omologo
Modello di riparazione di
DSB mediante
ricombinazione omologa
Ligazione dei
filamenti
2 giunzioni di
Holliday
La risoluzione del chiasma porta a:
Quali enzimi portano avanti la ricombinazione riparativa ?
La ricombinazione omologa in generale
Crossing-over meiotico
Trasfezione sperimentale di cellule con DNA ricombinante
Divisione cellulare:
http://www.cellsalive.com
/mitosis.htm
Only in meiosis
Crossing-over
(ricombinazione)
Ricombinazione sito-specifica
Nella ricombinazione degli elementi mobili
Caratteristica principale della ricombinazione omologa è che le due
molecole di DNA che ricombinano devono essere “omologhe”
ovvero
la loro sequenza deve avere un elevato grado di identità per
consentire lo scambio di filamenti e la migrazione per lunghi tratti.
trasposizioni, retrotrasposizioni, delezioni e inserzioni
dipedono da un tipo di ricombinazione diversa dalla ricombinazione
omologa
•
ricombinazione eterologa
•
ricombinazione sito-specifica
La ricombinazione sito-specifica:
CSSR (conservative site-specific recombination)
elemento mobile
brevi sequenze identiche
DNA ricevente
elemento mobile “integrato” nel DNA ricevente
Questo tipo di ricombinazione porta avanti diversi eventi:
inserzione
delezione
inversione
Le “ricombinasi” sono enzimi
che riconoscono corte sequenze
che si trovano, come ripetizioni
invertite, ai lati del punto di
inserzione
L’integrazione del fago lambda nel cromosoma di E. coli è un fenomeno
di ricombinazione sito specifica.
Lisogeno
Se il fago procede nella via lisogenica, si ha integrazione del DNA fagico nel
cromosoma di E. coli (ricombinazione sito-specifica).
The Human Genome Project
Hierarchical
Shotgun
The sequencing phase
Fragments are cloned into appropriate
vectors
Individual recombinants (bacterial clones) are grown, purified and sequenced (Sanger)
Il clonaggio classico
Sistema vettore-ospite in E. coli
Dideoxy-sequencing: the “chain-terminator” method of Sanger:
il metodo di maggior successo, si basa sulla sintesi di catene di DNA
complementari troncate in corrispondenza di una determinata base.
La sintesi avviene in presenza di molecole particolari: i di-deossi-nucleotidi
d
Reazione con di-deossi-Guanosintrifosfato
The label is usually 32P,
so that detection requires
autoradiography
Separazione con
elettroforesi
Una successiva evoluzione del metodo ha permesso l’automazione del
sequenziamento del DNA. In questo caso, i nucleotidi dideossiterminatori sono marcati mediante l’addizione di un gruppo chimico
fluorogenico, diverso per ogni base.
La reazione viene fatta insieme, ed ogni molecola “terminata” sarà
marcata col colore corrispondente alla base relativa.
Sequenziamento automatico del DNA
con dideossinucleotidi marcati con
fluorofori.
Le reazioni di terminazione di catena
vengono effettuate in una singola
provette, con ciascun
dideossinucleotide marcato con un
fluoroforo diverso.
Il frazionamento viene effettuato
con elettroforesi capillare.
Movie 1
Movie 2
Post-genome
1. Human genetic variation
1000 Human Genome project
exome project (exon-targeted resequencing)
2. Comparative genomic analysis
3. Functional analysis (ENCODE)
transcriptome
proteome
interactome
epigenome
…
Amplification of fragments
fragmentation
Adapter ligation
No cloning step
PCR
Library
No cloning step
NGS sequencing reads
Reads are mapped to the reference genome
reference genome
In re-sequencing, the number of independent sequences
(called «reads») is more important than lenght
The % of reference genome that is represented in «reads» is
the «coverage».
Other essential aspects:
1) speed
2) cost
3) error-to-depth ratio
Next Generation Sequencing
(deep-sequencing / mass sequencing)
generation of “DNA-nanoclones” on distinct solid surfaces by PCR or singlemolecule isolation
highly parallel in situ sequencing
record read-out i.e. millions or short sequences (“reads”)
align reads on genomes or assembly
Next generation sequencing methods:
Number of molecules per sequence
• Amplification
• Single-molecule
Biochemical measurement
• Sequencing by synthesis
(Sanger is synthesis + termination)
• Nucleotide chemistry
• Associated chemistry
• Sequencing by annealing and ligation
• Sequencing by direct physico-chemical measurements
Detection
• Optical detection
• Ion or conductance detection
Amplification by emulsion PCR (Roche 454, Polonator)
Biotinylated
template
http://www.pyrosequencing.com/DynPage.aspx?id=7454
In the second case, a surface sequencing is used
This is the 5’
Sequencing
direction
-CGCCTT
ATACGTCGTACTCGCAAGGCG
This must be 3’
This is the 3’
This is 5’
CGCCTT-
AAGGCGTACGTCGTACTCGCAA
Sequencing
direction
CGCCTT-
AAGGCGTACGTCGTATCTCGCAA
TTGCGA
This is the 3’
Sequencing
direction
Il vetrino viene chiuso per dare una camera microfluidica
Le frecce indicano il flusso di reagenti chimici necessari alle reazioni che
rivelano progressivamente l’ordine dei nucleotidi di ciascun frammento
Un laser scanner registra in continuo la fluorescenza in tutto il
vetrino, ogni segnale corrisponde ad una «base» (es. Red=A,
green=T, yellow=G, blue=C ) leggendo così tutte le sequenze
Reading: 2 spots are exemplified here:
Immagine monocolore: sono esguiti per ogni posizioe quattro cicli
successivi, con ciascuna delle basi (come reversible terminator
Il «readout» sono milioni di corte sequenze (secondo i tipi, da 30 a 500 nucleotidi)
che vengono chiamate «reads»
Tipicamente, ogni campione produce tra 10 e 100 milioni di «reads»
I «reads» vengono quindi allineati al genoma e assegnati di conseguenza ai diversi
geni.
sequence
# of reads map
region
CTAGTCATGCTCTCGATCGGTCATAGTTTAGTCTGACT
12
Chr 1: 12,345,678-12,345,711
CHD1 coding
TTAAAGTACTGTCATGATTTCATGCTAGCTTTTCAAAG
3
Chr 7: 1,987,654-1,987,694
PTT1 intron
GGCTACTAGTCTATTACAAGGGCATCGCGGATCGCGT
1
Chr 16: 23,456,711-23,456,755
intergenic
22
Chr 1: 12,345,123-12,345,183
CHD1 coding
....
....
TACTGCTGACGCCGCATGCATTTACGCTGCGGCATCGG
....
Illumina-Solexa Genome Analyzer
Read lengths: 36 bp, 50 bp, 75 bp, 100bp for fragment or paired-end sequencing
Throughput (reads): 120 million reads per run, fragment
SoliD – Applied Biosystems
Read lengths: 50 bp fragment, 25 bp and 35 bp paired
Throughput (reads): > 160 million reads per slide, fragment
Roche – 454
Read lengths: Averaging 350 - 400 bp
Throughput (reads): ~1 million reads per run
Extensive sequencing of RNA from several cell types and tissues has provided
answer to a long-lasting question:
Why do Humans have so few genes ?
To be sequenced, RNA must be copied to DNA (called cDNA – complementary DNA)
This is done using an enzyme called Reverse Transcriptase (RT)
cDNA is then fragmented, linked to adaptors and used to generate libraries for NGS
Wang et al. (2009). Nat. Rev. Genet. 10:57-69
Wang et al. (2009)
Figure 1 | A typical RnA-seq
experiment. Briefly, long
RNAs are first converted into
a library of cDNA
fragments through either
RNA fragmentation or DNA
fragmentation (see main
text). Sequencing
adaptors (blue) are
subsequently added to each
cDNA fragment and a short
sequence is obtained from
each cDNA using highthroughput sequencing
technology. The resulting
sequence reads are aligned
with the reference genome or
transcriptome, and classified
as three types: exonic reads,
junction reads
and poly(A) end-reads. These
three types are used to
generate a base-resolution
expression profile for
each gene, as illustrated at
the bottom; a yeast ORF with
one intron is shown.
Information is dependent on the kind of RNA preparation. Due to the uneven
representation of each RNA species, it is unpractical to run RNA-Seq analysis using
«total RNA preparations» (most of the reads would map to highly represented RNAs,
whereas rare RNA wouldn’t be represented at all in the sequencing library)
Cells, tissue
Tot RNA
fractionation
By type:
non-rRNA
all nonribosomal
By size:
Small < 200 nt
Long > 200 nt
small noncoding
coding + other
By adds:
poly(A)+
poly(A)-
>99% coding
large number noncoding
noncoding
By location:
nuclear
cytoplasmatic
snRNA, snoRNA, lncRNA
coding, noncoding
By function:
exome
microarray-selected
coding exons
guanine-N7methyltransferase
Elongation
factor
EJC deposited 20-24 nt upstream the
exon-exon junction
Splicing factors and snRNPs are replaced by the
EJC (exon junction complex) proteins
From Aguilera 2005, Curr Op Cell Biol, 17:242.
How prevalent is splicing (i.e. exon-intron gene organization)
in different organisms ?
S. cerevisiae
has only 253 introns (3% of genes),
only 6 genes have 2 introns.
S. pombe
43% of the genes have introns, many
of them contains >1 intron
H. sapiens
>99% of genes contain multiple introns
(40-75 nt)
Average human gene:
Length: 28,000 bp
No. of exons: 8.8
Exon length: 120 bp
No. of introns: 7.8
Intron length: 10 to >100.000 bp
Da «intronless» a pochi introni, a
parecchie decine
one intron in the human neurexin gene is approx. 480,000 nt !
Lo splicing alternativo consiste nella scelta di considerare esoni o
introni alcune sequenze.
Storico: 1983
Un gene umano va incontro a
processamento differenziale
La calcitonina è un ormone costituito
da un polipeptide di 32-aminoacidi che
viene prodotto, negli esseri umani,
dalle cellule parafollicolari della tiroide.
La principale funzione della calcitonina
è l'abbassamento della concentrazione
di calcio nel sangue
CGRP is produced in both peripheral
and central neurons. It is a potent
peptide vasodilator and can function in
the transmission of pain. In the spinal
cord, the function and expression of
CGRP may differ depending on the
location of synthesis.
Alternative Splicing
may concern one or
more exons.
Quite often many
isoforms are coexpressed;
sometimes there are
tissue-specific
isoforms.
Alternative Splicing of fibronectin pre-mRNA
Introns are drawn not to scale
Alternative splicing
H. sapiens
S. cerevisiae:
H. sapiens:
Estimated number of protein-coding genes: < 22,000
Estimated number of proteins: > 90,000
253 genes contain introns
only 3 genes shown experimentally to undergo alternative splicing
>99% predicted to have exon-intron structure
>90% predicted to undergo alternative splicing
Materiale per uso didattico
AS in many cases give rise to proteins with differential functions and roles.
One example is already well-known in this course: ERBB4
(e) Isoforms of the Slo protein lacking sequences encoded by the
STREX exon have fast deactivation kinetics and low Ca 2+ sensitivity,
whereas isoforms containing STREX-encoded sequences have slower
deactivation kinetics and higher Ca 2+ sensitivity.
From: Graveley BR (2001) Trends Genet., 17:100-106.
Fig. 1. Alternative splicing of the slo gene.
(a) The mammalian cochlea. The cochlea
is a snail-like structure of the inner ear
that contains hair cells organized along a
basilar membrane. The basilar membrane
traverses the length of the curled-up
cochlea.
(b) The cochlea is sliced transversely as
shown in (a) and the section of the
cochlea containing the basilar membrane
and the hair cells depicted. There are four
rows of hair cells, one inner hair cell and
three outer hair cells, situated above the
basilar membrane.
(c) The cochlea is unrolled to reveal the
basilar membrane viewed from above. The
four hair cells are arranged in rows along
the length of the basilar membrane. The
hair cells are tuned to unique narrow
sound frequencies along the basilar
membrane creating a tonotopic gradient.
At one end of the membrane, hair cells
are tuned to respond to a frequency of 20
Hz, where as hair cells at the other end
respond to 20 000 Hz.
(d) Organization of the human slo gene.
The exon–intron organization of the
slogene (determined by an analysis of
draft sequence of the human genome) is
depicted. The constitutive splicing events
are indicated below the gene and
alternative splicing events are depicted
above the gene. The constitutive exons
are white and the alternative exons are
shaded. The STREX exon is purple.
present in the postsynaptic cell, and thus function to initiate synaptogenesis. In contrast, b-neurexin I containing exon 20 encoded sequences can not
interact with neuroligins. This form of b-neurexin I might indirectly function in releasing synapses.
Neurexins
From: Graveley BR (2001) Trends Genet., 17:100-106.
Drosophila Dscam gene provides probably the extreme example of
alternative splicing.
Perhaps the most complex event that takes place during development
is the migration and connection of neurons. Even in a ‘simple’ organism
such as Drosophila melanogaster, which contains only ~250 000
neurons, accurately wiring neurons together would appear to be a
daunting task.
In flies, the gene encoding the Down syndrome cell adhesion molecule
(Dscam) appears to fulfill at least part of this role. Dscam encodes an
axon guidance receptor with an extracellular domain that contains ten
immunoglobulin (Ig) repeats. The most striking feature of the Dscam
gene is that it’s pre-mRNA can be alternatively spliced into over
38,000 different mRNA isoforms (Fig. 3a). This is 2–3 times the
number of predicted genes in the entire organism !
Each mRNA encodes a distinct receptor with the potential ability to
interact with different molecular guidance cues, directing the growing
axon to its proper location.
as an axon guidance receptor. It is thought that each Dscam variant will interact with a unique set of axon guidance cues.
The form of Dscam shown on the left will interact with guidance cue A. The form of Dscam shown on the right contains different sequences encoded by exons
4, 6 and 9 and thus interacts with guidance cue B, rather than guidance cue A. Neurons expressing the form of Dscam shown on the right will be attracted in a
different direction than neurons expressing the form shown on the left.
From: Graveley BR (2001) Trends Genet., 17:100-106.
Potentially 38,000 splicing variants
How extensive is Alternative Splicing usage in Humans ?
Exon-exon junction micro-arrays
Oligonucleotide probes, typically 25–60 nucleotides in length, can be designed to hybridize to isoform-specific mRNA regions.
Recently, alternative splicing microarrays have been designed with probes that are specific to both exons and exon–exon junctions.
Probes e1, e2 and e3 are exon specific, whereas j1–2, j2–3 and j1–3 are isoform-specific junction probes. Some arrays also
contain intron probes (i1 and i2) to indicate signals from pre-mRNA. Various array design and data processing strategies facilitate
the quantitative analysis of alternative splicing patterns, some of which have been subsequently confirmed by PCR after reverse
transcription of RNA (RT-PCR). Johnson et al.(2003) used arrays with probes for all adjacent exon–exon junctions in 10,000 human
genes and hybridized these with samples from 52 human tissues and cell lines. This revealed cell-type-specific clustering of
alternative splicing events, and allowed the discovery of new alternative splicing events. Pan et al. analysed 3,126 known cassettetype alternative splicing events in mouse using exon-specific and exon–exon junction probes. Analysis of RNAs in ten tissues
showed clustering of alternative splicing events by tissue type, and further revealed that tissue-specific programmes of transcription
and alternative splicing operate on different subsets of genes. A direct comparison also showed that computational prediction of
tissue-specific alternative splicing based on ESTs and cDNAs performed poorly compared with the alternative splicing microarray
and RT-PCR.
From: Matlin et al. (2005), Nature Rev Mol Cell Biol, 6: 386.
RNA-Seq (NGS)
Figure 1 | Frequency and relative abundance of alternative splicing isoforms in human
genes.
a, mRNA-Seq reads mapping to a portion of the SLC25A3 gene locus. The number of
mapped reads starting at each nucleotide position is displayed (log10) for the tissues listed
at the right. Arcs represent junctions detected by splice junction reads.
Bottom: exon/intron structures of representative transcripts containing mutually exclusive
exons 3A and 3B (GenBank accession numbers shown at the right).
b, Mean fraction of multi-exon genes with detected alternative splicing in bins of 500
genes, grouped by total read count per gene. A gene was considered as alternatively
spliced if splice junction reads joining the same 5’ splice site (5’SS) to different 3’ splice
sites (3’SS) (with at least two independently mapping reads supporting each junction), or
joining the same 3’SS to different 5’SS, were observed. The true extent of alternative
splicing was estimated from the upper asymptote of the best-fit sigmoid curve (red
curve). Circles show the fraction of alternatively spliced genes.
«Pure» alternative splicing
Figure 3
Types of alternative splicing.
In all five examples of alternative
splicing, constitutive exons are
shown in red and alternatively
spliced regions in green, introns are
represented by solid lines, and
dashed lines indicate splicing
activities. Relative abundance of
alternative splicing events that are
conserved between human and
mouse transcriptomes are shown
above each example (in % of total
alternative splicing events).
From: Ast G. (2004)
Nature Rev Genetics 5: 773.
Note that the indicated percentages derive from
older studies and are slightly different from
those demonstrated by recent, RNA-Seq based
evaluations
Figure 2 | Pervasive tissue-specific regulation of alternative mRNA isoforms. Rows represent the eight
different alternative transcript event types diagrammed. Mapped reads supporting expression of upper
isoform, lower isoform or both isoforms are shown in blue, red and grey, respectively. Columns 1–4
show the numbers of events of each type: (1) supported by cDNA and/or EST data; (2) with ≥ 1 isoform
supported by mRNA-Seq reads; (3) with both isoforms supported by reads; and (4) events detected as
tissue regulated (Fisher’s exact test) at an FDR of 5% (assuming negligible technical variation).
Columns 5 and 6 show: (5) the observed percentage of events with both isoforms detected that were
observed to be tissue-regulated; and (6) the estimated true percentage of tissue-regulated isoforms
after correction for power to detect tissue bias (Supplementary Fig. 6) and for the FDR. For some
event types, ‘common reads’ (grey bars) were used in lieu of (for tandem 39UTR events) or in
addition to ‘exclusion’ reads for detection of changes in isoform levels between tissues.
Note that Aa use the following definition for “tissue-specific”: at least 10% variation in isoforms.
Il progetto ENCODE ha mostrato che la
maggior parte dei geni umani dà
origine a trascritti plurimi, utilizzando:
• Siti di inizio alternativi
• Ampie variazioni nel quadro di
splicing
• 3’ alternativi
Il numero medio tendenziale sarebbe
tra 9 e 12 trascritti.
Figure 1 from Licatalosi et al., 2010
Some genes display “alternative promoters”
Proximal
promoter
Distal
promoter
5’
3’
1
2
Sometimes an exon is present between the two promoters
alternative parts
5’
3’
1
2
Coding or
noncoding
exon
If an acceptor site and a donor site are present
This is different from the story of multiple TSS
Unique TSS
5’
5’
3’
1
3’
1
Multiple TSS
Other genes possess “alternative polyadenylation sites”
Distal pA
site
Proximal pA
site
5’
3’
1
stop
stop
Coding or
noncoding
exon
How is Alternative Splicing Regulated ??
Costitutivo:
snRNP (U1,U2)
Siti di splicing
«conservati»
Alternativo:
elementi cis
(ESE, ESS, ISE,
ISS) riconosciuti
da SR, hnRNP,
Attivatori,
Repressori
Basics of the mechanisms of alternative splicing. (a) The architecture of a pre-mRNA and the important cis-acting
sequence elements that direct the splicing reaction. The consensus sequences for the 50 splice site, branchpoint and 3’
splice site for human introns is shown. (b) Schematic diagram of the sequences and proteins involved in regulating
alternative splicing. Four types of regulatory sequences are known: intronic splicing enhancers (ISEs), intronic splicing
silencers (ISSs), exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs). The enhancer elements are
recognized by activator proteins. Within exons, these activators are most commonly members of the SR protein family.
The silencer elements are bound by repressor proteins. Within exons, these repressors tend to be members of the
hnRNP protein family. Regardless of their binding location, activators tend to enhance the binding of spliceosomal
components to the regulated splice site while repressors tend to inhibit binding or function of the spliceosomal
components.
From: McManus & Graveley, COGD, 2011
Pervasive Transcriptome
RNA-Seq is used both for quantitative and qualitative evaluation of transcriptomes
Read mapping
Quantitative (measuring gene expression)
In this case, reads are mapped to a reference «transcriptome»,
then the number of reads for each known RNA is counted to
obtain a table of expression or a density graph.
Qualitative:
discovering
• new RNA transcripts
• new exon-exon junctions (i.e. alternative splicing forms)
• aberrant transcripts and splicing forms
•new RNA forms (e.g. circular RNA)
Experiments published to date using RNA-Seq
show extensive «pervasive» transcription of genomes
with more than 20,000 NONCODING RNA transcripts (human)
Noncoding are high percentage qualitatively
low percentage quantitatively
Nota: i numeri sono un po’
diversi da quelli che
successivamente presenterà
ENCODE. La differenza è
dovuta al numero di
esperimenti considerati, che
qui sono relativamente pochi
(2007-2008).
Ponting & Belgard, 2009
ENCODE
Today: many RNA-Seq experiments on tissues, cell lines published.
ENCODE reports results from 15 cell lines
RNA-Seq from either «cytoplasmic» or «nuclear» fractions, poly(A+) or (A-) RNA
62% of genomic bases are represented in long RNA transcripts (>200nt)
only 5.5% map to GENECODE exons
only 31% were classified «intergenic»
CAGE analysis: 62,403 TSS
Nuclear to cytoplasmatic analysis showed many transcripts processed
to give short RNA (<200)
Very elevated prevalence of Alternative splicing
Each gene show several transcripts (6-9 per gene, depending on
classification) plateauing at 12.
Short noncoding
snRNA
small U-RNA components of spliceosome particles
snoRNA
small RNA that guide chemical modifications to other RNAs
micro-RNA (miRNA)
single-stranded 21-24 nt post-transcriptional regulators
siRNA
double-stranded 21-25 bp , silencing RNA various pathways
piRNA
26-31 nt involved in transcriptional silencing (transposons)
sRNA
various transcription associated small RNA, unknown
Long noncoding (15-20,000 transcripts identified)
The simplest classification of long ncRNAs is based on their loci of origin.
• Antisense transcripts
Transcribed in antisense of coding RNA
• Intronic transcripts (sense or a/s)
Spanning introns
• Divergent transcripts
starting from promoters in opposite
direction
• Long intergenic transcripts
Transcript in gene-desert regions
• e-RNA
In either direction, starting from active
enhancers
No class-specific function identified.
Sporadic functions described:
• Transcriptional regulators, connecting activators or repressors
• Silencing, by acting as scaffold for chromatin modifying enzymes
• «Sponges» for micro RNA
• Translational regulators
Dal punto di vista della regolazione, le due classi più nuove e
più interessanti sono:
micro RNA
lnc RNA
Nella seconda parte di regolazione

![mutazioni genetiche [al DNA] effetti evolutivi [fetali] effetti tardivi](http://s1.studylibit.com/store/data/004205334_1-d8ada56ee9f5184276979f04a9a248a9-300x300.png)