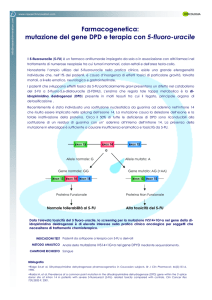
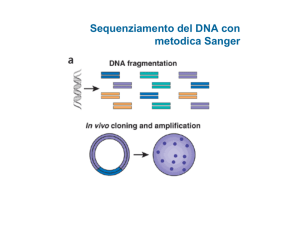
Tecniche per la produzione di dati genomici
NGS Illumina sequencing:
a pratical overview
Federica Cattonaro
Istituto di Genomica Applicata
IGA Technology Services Srl
(Udine, Italy)
Convegno ‘La genomica al servizio dell’agricoltura’
Università Cattolica del Sacro Cuore
Piacenza, 25 maggio 2012
Illumina target enrichment protocol
TARGET ENRICHMENT OPTIONS
Custom target
You can choose which region and for which organisms (need a reference) to do
genome capture
Region captured from 200Kb to 32Mbp for Agilent SureSelect system
Region captured from 700Kb to 15Mbp for Illumina system
Software to design the probes: Illumina Design Studio, Agilent eArray
ONLY human target resequencing
Human all exon
Agilent SureSelect all exon 50Mbp= target size about 50 Mbp
Illumina TrueSeq exon enrichment kit = target size about 62Mbp
Custom target resequencing
Amplicon Seq (Illumina): 12 to 96 Kbp
Halopex (Agilent): until 1Mbp
SureSelect All Exon – Towards non-Human Species
All Exon Mouse:
49.6Mb design. Exon definition derived from Ensembl + RefSeq,
designed against mm9 reference from UCSC
All Exon Bovine:
64Mb design. The library is based on University of Maryland build 3.1
against NCBI exons as well as predicted exons, UTRs, and miRNAs.
Worked with USDA on design.
All Exon Canine:
54Mb design. UCSC tracks from CanFam2 for ensembl as well as
Refseq, human protein alignments, and spliced ESTs that lie outside of
ensembl.
All Exon Zebrafish:
96Mb design. Zv9 library is in Emsembl and is annotated with gene and
exon coordinates. Worked with Sanger on design.
Note: All Exon Bovine, Canine and Zebrafish are being provided in Early Access
Format
1. Illumina TruSeq DNA Sample prep bisulfite modified
(C to U changes in non metylated cytosine)
1. ME-DIP analysis (antibody vs methylated C)
Bioanalyzer, Qbit fluorimeter, Real-Time....
IGA Technology Services S.r.l.
(commercial spin-off of IGA)
NGS facilities
Illumina Genome Analyzer IIx (April 2010): 95 Gbp/run (pair-end 2x150bp)
(already dismissed!)
HiSeq2000 (by December 2010): 600 Gbp/run (pair-end 2x100bp)
MiSeq (by April 2012): 2 Gbp/run (pair-end 2x150bp)
Esempi di servizi alla ricerca legati al
sequenziamento NGS
applicazione
attività
Metagenomica
sequenziamento NGS di comunità ambientali (16S o tutte le
molecoile di RNA/DNA)
Analisi di espressione
genica
sequenziamento NGS dell’mRNA (trascrittoma) per l’analisi
dell’espressione genica in specie vegetali, in bovini, cani, uomo...
Analisi di smallRNA
annotazione e quantificazione
Ricostruzione
trascrittomi
sequenziamento NGS per la ricostruzione delle sequenze codificanti
(geni) in varie specie il cui genoma non è stato sequenziato, come
tasso, abete bianco, anemone di mare, crostacei...
Sequenziamento di
piccoli genomi
sequenziamento NGS di ceppi batterici
Identificazione
varianti SNP e INDELs
Sequenziamento esomi umani, tumori (controllo vs tumore), target
resequencing