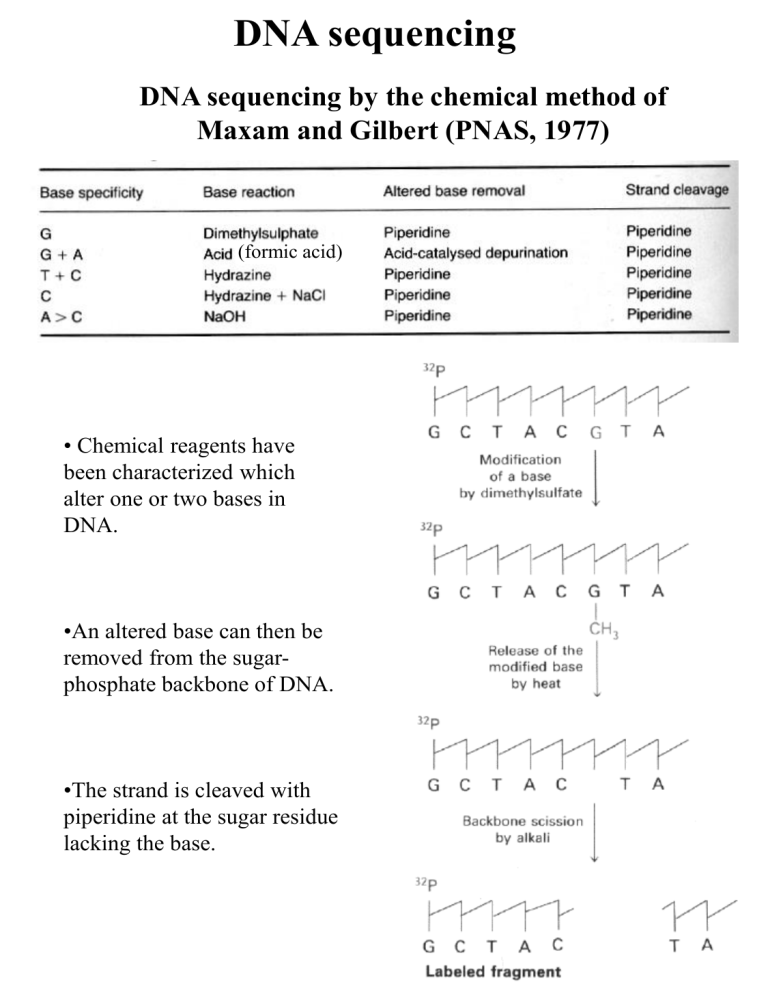
DNA sequencing
DNA sequencing by the chemical method of
Maxam and Gilbert (PNAS, 1977)
(formic acid)
• Chemical reagents have
been characterized which
alter one or two bases in
DNA.
•An altered base can then be
removed from the sugarphosphate backbone of DNA.
•The strand is cleaved with
piperidine at the sugar residue
lacking the base.
Reading the DNA sequence
Gel PAGE + Urea (6 M)
Sequencing by the chain-terminator or
dideoxy procedure (Sanger, 1977)
- Enzymatic methods.
- Random incorporation of a dideoxynucleoside
triphosphate into a growing strand of DNA.
This method is an in-vitro DNA synthesis
using ‘terminators’. Incorporation of dideoxynucleotides into growing strand
terminates synthesis.
- Requires DNA polymerase I. Requires a
cloning vector with initial primer (M13, high
Nobel winner 1980
yield bacteriophage).
- Uses 32P-deoxynucleoside triphosphates.
-Synthesized strand sizes are determined for each di-deoxynucleotide by
using gel or capillary electrophoresis.
Principle of the method
3’
5’
T
primer
TT
T
3’
5’
ddATP in the
reaction: anywhere
there’s a T in the
template strand,
occasionally a ddA
will be added to the
growing strand
ddA
ddA
ddA
ddA
The dideoxy chain termination (or enzymatic) method of DNA
sequencing involves the in vitro synthesis of a DNA strand by a
DNA polymerase, such as:
Klenow fragment of E.coli DNA polymerase I (used in
combination with cloning the DNA to be sequenced in M13 series
of single-stranded vectors);
modified form of phage T7 DNA polymerase, Sequenase.
This enzyme, developed by Tabor and Richardson (P.N.A.S., 1987,
vol. 84:4767-4772) is a site-directed mutant (His123Glu) of
bacteriophage gene 5 protein.
Features of Sequenase:
1. unlike Klenow fragment, Sequenase can be used with of
double-stranded vectors);
2. reduced exonuclease activity,
3. highly processive; catalyzing the polymerization of thousands of
nucleotides without dissociating from the template.
Taq DNA polymerase (used in cycle sequencing - PCR)
Primer walking
La reazione di sequenza permette di stabilire con buona certezza l’ordine
dei primi 250-350 nucleotidi. Gli inserti di DNA clonati sono solitamente
molto più lunghi (5000 bp).
Determinata la sequenza del primo tratto, si sintetizza un secondo primer
disegnato per ibridarsi con la regione lontana circa 300 basi a valle del
sito di innesco del primo primer.
In maniera simile si sceglie un terzo sito legame per l’innesco, si
sintetizza un altro oligonucleotide e si determina la sequenza delle
successive 250-350 basi. La strategia “Primer walking” va avanti fino a
completare il sequenziamento dell’intero inserto.
Enzymatic
method
Chemical
method
Un apparato per il
sequenziamento su
gel di poliacrilamide
sottile ~ 0.2-0.4 mm.
MAXAM & GILBERT
METHOD
SANGER
METHOD
by-pass all the problems
associated with polymerases
RAPID; a large n° of
samples can be
processed
simultaneously
does not require subcloning into
seq. vectors (restriction fragments
can be used directly)
composition of 2-D
structure of the DNA
template can cause
premature termination
by DNA polymerase
time-consuming (labeling of a
single end, purification steps)
the only method for sequencing
small oligonucleotides
background due to degradation
Corsa
breve
Corsa
lunga
Automated DNA Sequencing
These systems employ fluorescent
dyes attached to either the primer (I°
generation of this techniques) or the
ddNTP (II° generation of this techniques).
The DNA fragments produced by
sequencing reactions are run through
polyacrylamide gels or capillary
electrophoresis.
The detection systems relies on laserinduced fluorescence (helium-neon laser;
633 nm).
Detecting the bands within the gel is not trivial as there are only
about 10-15 to 10-16 moles (femtomoles) of DNA in each band.
(laser)
Proc. Natl. Acad. Sci. USA (1995)
vol.92, pp.4347-4351
Four-dye primer sequencing is one of the most commonly used method
for high-throughput DNA sequencing. As in other sequencing
methodologies, the detection sensitivity is limited by the spectroscopic
properties of the available dyes (based on the structure of fluorescein) for
labeling the sequencing fragments.
Structure of FLUORESCEIN and
FAM (5-carboxyfluorescein)
To optimize the absorption and emission properties of the label,
primers have been developed that exploit fluorescence energy
transfer (ET)
Fluorescence ET is mediated by a dipole-dipole coupling between
two chromophores that results in resonance transfer of excitation
energy from an excited donor molecule to an acceptor.
Amplified
signal
D
A
ET primers have two fluorescent dyes attached. The effective
fluorescence intensity is 2 to 10 times greater than single dye
primers. FAM is selected as common donor, FAM, JOE, TAMRA
and ROX are selected as acceptors.
FAM
5-carboxyfluorescein
(SE= Succinimidyl ester)
JOE
2’,7’-dimethoxy-4’,5’-dichloro-
-6-carboxyfluorescein
R = -COOH
TAMRA
ROX
tetramethyl-6-carboxyRhodamine
6-carboxy-X-Rhodamine
R1 = H
R1 = H
R2 = -COOH
R2 = -COOH
A standard procedure
(II° generation sequencing)
1)
2)
3)
The DNA is prepared as
single strand
A mixture of four normal
(deoxy) nucleotides
(dGTP, dATP, dTTP,
dCTP)
A mixture of four
dideoxynucleotides (each
present in limiting
amounts) each labeled
with a tag that fluoresces
a different colour
(ddGTP, ddATP, ddTTP,
ddCTP)
4)
DNA polymerase
5)
Adequate buffer
Results can be monitored in real-time on the interfaced screen and
subsequently subjected to graphically interactive analysis
READ LENGTHS:
home-made PAGE apparatus ( 17 X 36 cm. - 0.3 mm thick gel)
up to 150 - 180 bp;
Macrophor Electrophoresis Unit (patented design of the EMBL)
LKB-Pharmacia. 20 X 50 cm. - 0.1 mm thick gel. The
electrophoresis unit is equipped with a thermostatic plate that
provides uniform temperature control (eliminates ‘smiling effects’
and resolves G-C compressions) up to 300 - 400 bp;
ALF DNA Sequencer Equipped with fixed-laser detection
system, scanning a polyacrylamide gel (Pharmacia) up to
500 bp/hour/lane
ABI Prism 3700 DNA Analyzer (Applied Biosystem). Automated
capillary gel electrophoresis system. All four sequencing
reactions are run in a single capillary (dye-labeled terminator
chemistry). Detect over 500 bases at 98.5% accuracy at 100
bases/hour/capillary.
MegaBACE (Amersham-Pharmacia Biotech). DNA fragments are
separated by capillary electrophoresis (16, 48 or 96-capillary).
It is operated by a confocal scanning laser, and is capable of up
to 12 DNA sequencing runs per 24-hour (read length >650 bp),
producing up to 500.000 bases/day.
Next Generation Sequencing
Pirosequenziamento
Ronaghi M, Ehleen M and Nyrén P (1998) A sequencing method based on
realtime pyrophosphate. Science, 238, 363-365.
Si basa sulla rilevazione del
pirofosfato rilasciato
dall’incorporazione di un
nucleotide durante la sintesi del
DNA.
adenosine 5’phosphosulfate
(APS)
Apyrase is an ATP
diphosphohydrolase. It
catalyses the removal of
the gamma phosphate
from ATP and the beta
phosphate from ADP.
The phosphate from
AMP is not removed.
PPi is not
produced
• Il primer è ibridato allo stampo a singolo elica, amplificato per PCR, e
incubato con gli enzimi DNA polimerasi, ATP sulfurilasi, luciferasi e
apirasi, adenosin 5’ fosfosolfato (APS) e luciferina.
• Il primo dei quattro dNTP viene aggiunto alla reazione. La DNA
polimerasi catalizza l’incorporazione del dNTP al filamento di DNA, se è
complementare alla base del filamento stampo.
Ogni evento di incorporazione è accompagnato dal rilascio di piro-fosfato
(PPi) in quantità equimolare a quella del nucleotide incorporato.
• In presenza di adenosina 5’ fosfosolfato (APS), l’ATP sulforilasi
converte quantitativamente il PPi ad ATP, che, a sua volta guida
la conversione, catalizzata dalla luciferasi, di luciferina ad
ossiluciferina con conseguente produzione di luce di intensità
proporzionale alla quantità di ATP.
La luce prodotta è rilevata da una CCD camera e visualizzato
come picco in un pirogramma.
• L’apirasi, un enzima che degrada nucleotidi, degrada continuamente tutti
i dNTP non incorporati e l’ATP in eccesso. L’apirasi non produce PPi. Non
appena la degradazione è completata viene aggiunto un altro dNTP.
• I dNTP vengono aggiunti
sequenzialmente, uno alla volta. Poiché il
dATP è un substrato naturale della
luciferasi (come la ATP), al suo posto viene
utilizzato la deossiadenosina
α-tio-trifosfato (dATPS) che viene utilizzata
efficentemente dalla DNA polimerasi ma
non viene riconosciuta dalla luciferasi.
• Man mano che il processo continua,
il filamento di DNA complementare è
sintetizzato e la sequenza
nucleotidica è determinata dai picchi
del pirogramma (~ 300 basi).
La sequenza è: TTTGGGGTTGCAGTT →
+ DNA polimerasi, apirasi +
ATP sulforilasi + luciferasi
454 Technology (Roche)
•
To start, the DNA is sheared
into 300-800 bp fragments, and
the ends are “polished” by
removing any unpaired bases at
the ends.
•
Adapters are added to each
end. The DNA is made single
stranded at this point.
•
One adapter contains biotin,
which binds to a streptavidincoated bead. The ratio of beads
to DNA molecules is controlled
so that most beads get only a
single DNA attached to them.
•
Oil is added to the beads and an
emulsion is created. PCR is
then performed, with each
aqueous droplet forming its
own micro-reactor. Each bead
ends up coated with about a
million identical copies of the
original DNA.
Biotinylated primers
•
After the emulsion PCR has been performed,
the oil is removed, and the beads are put
into a “picotiter” plate. Each well is just
big enough to hold a single bead.
•
The pyrosequencing enzymes are attached
to much smaller beads, which are then
added to each well.
•
The plate is then repeatedly washed with the
each of the four dNTPs, plus other necessary
reagents, in a repeating cycle.
•
The plate is coupled to a fiber optic chip.
A CCD camera records the light flashes
from each well.
Illumina/Solexa Sequencing
- This method uses the basic Sanger idea of “sequencing by
synthesis” of the second strand of a DNA molecule. Starting
with a primer, new bases are added one at a time, with
fluorescent tags used to determine which base was added.
- The fluorescent tags block the 3’-OH of the new
nucleotide, and so the next base can only be added when
the tag is removed.
-So, unlike pyrosequencing, you never have to worry about
how many adjacent bases of the same type are present.
-The cycle is repeated 50-100 times.
The idea is to put 2 different adapters on each end of the DNA, then
bind it to a slide coated with the complementary sequences for each
primer. This allows “bridge PCR”, producing a small spot of amplified
DNA on the slide. The slide contains millions of individual DNA
spots. The spots are visualized during the sequencing run, using
the fluorescence of the nucleotide being added.
Attached
termini
Template DNA
●●●●●
●● ●●●
●
PCR Product
Third generation sequencing
Back in 2003, The Human Genome cost approximately $500 million, years
of work and huge international effort to produce. Actually, the cost of a
genome falls to just $10,000 and maybe as low as $1000.
Genome Sequencer FLX
(Roche analyzer)
Illumina Genome Analyzer
Roche 454 FLX
Illumina Genome
Analyzer
Amount of starting
material needed
DNA: 3 to 5 μg
Total RNA: 20 μg
DNA: 1 to 5 μg
Total RNA: 1 to 2 μg
Sequencing
technology
Pyrosequencing
Bridge amplification
Read length
200-300 bases
25-35 bases
Sequence yield
100Mb
800Mb-2Gb
Data file
12 to 15Gb/run
1 Tbyte
Time/run
8hrs
3 to 5 days


![mutazioni genetiche [al DNA] effetti evolutivi [fetali] effetti tardivi](http://s1.studylibit.com/store/data/004205334_1-d8ada56ee9f5184276979f04a9a248a9-300x300.png)
![ESTRAZIONE DNA DI BANANA [modalità compatibilità]](http://s1.studylibit.com/store/data/004790261_1-44f24ac2746d75210371d06017fe0828-300x300.png)



![(Microsoft PowerPoint - PCR.ppt [modalit\340 compatibilit\340])](http://s1.studylibit.com/store/data/001402582_1-53c8daabdc15032b8943ee23f0a14a13-300x300.png)


